- Title
-
Role of Ribosomal RNA Released from Red Cells in Blood Coagulation in Zebrafish and Humans
- Authors
- Alharbi, A., Iyer, N., AlQaryoute, A., Raman, R., Burks, D., Azad, R., Jagadeeswaran, P.
- Source
- Full text @ Blood Adv
|
|
|
|
|
|
|
|
|
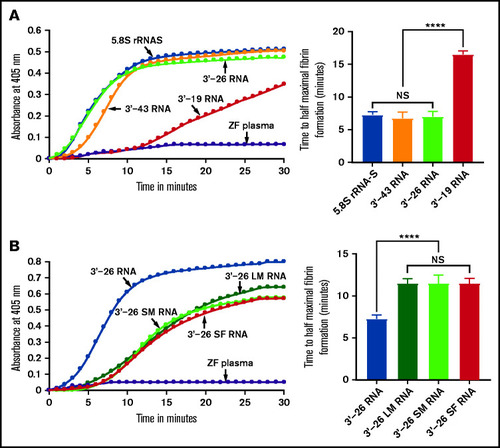
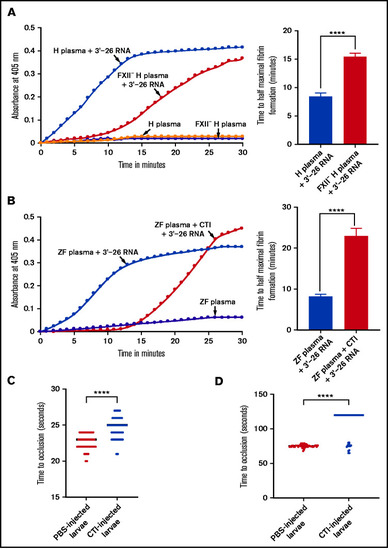
Lack of activation of FXII-deficient human plasma or zebrafish plasma in the presence of CTI by 3'-26 RNA. (A-B) Left graphs show the increasing fibrin formation with increasing time in the kinetic coagulation assay. (A) Left graph: curves were obtained from normal and FXII-deficient human plasma in the presence of 3'-26 RNA and in the absence of RNA. Bar graph shows a significant shortening of the time to half-maximal fibrin formation (using data obtained from the left graph); normal human plasma is compared with FXII-deficient human plasma in the presence of 3'-26 RNA. (B) Left graph: curves were obtained from zebrafish plasma activated by 3'-26 RNA in the absence or presence of CTI and in the absence of both RNA and CTI. Bar graph shows a significant shortening of the time to half-maximal fibrin formation (using data obtained from the left graph) for zebrafish plasma activated by 3'-26 RNA in the absence or presence of CTI (n = 4). For curves in panels A and B, the time in minutes was plotted against the absorbance at 405 nm at 25°C. The data for time to half-maximal fibrin formation (in minutes) were analyzed using Student t test and are shown as mean ± SEM. (C-D) Inhibition of venous thrombosis (C) and arterial thrombosis (D) by CTI in zebrafish larvae. Prolongation of the TTO in the caudal vein (C) and the caudal artery (D) of CTI-injected larvae was significant compared with that for the PBS-injected larvae (n = 48). The data were analyzed using Student t test and are shown as mean ± SEM. ****P < .0001. |
|
|
|
|