- Title
-
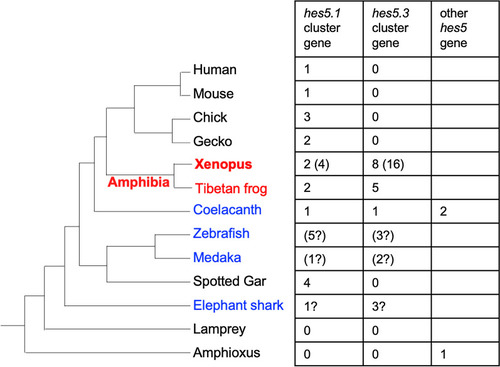
Evolution of hes gene family in vertebrates: the hes5 cluster genes have specifically increased in frogs
- Authors
- Kuretani, A., Yamamoto, T., Taira, M., Michiue, T.
- Source
- Full text @ BMC Ecol Evol
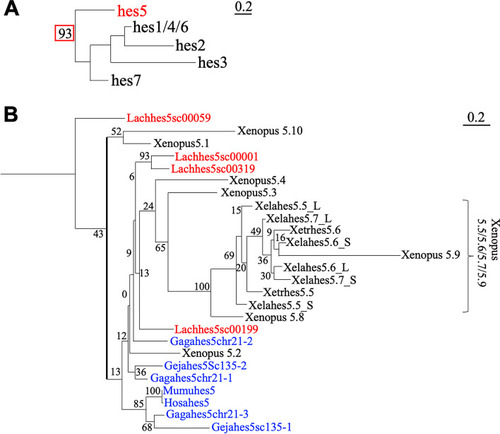
|
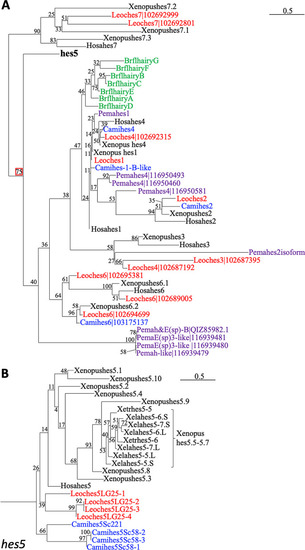
Phylogenetic analysis of |
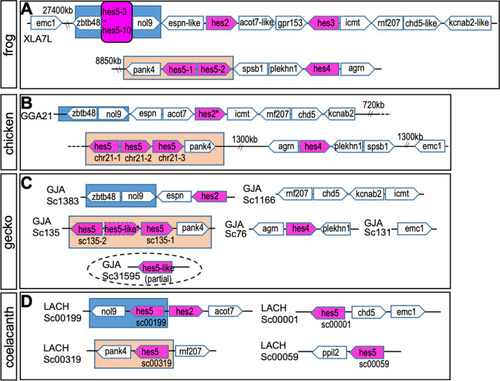
|
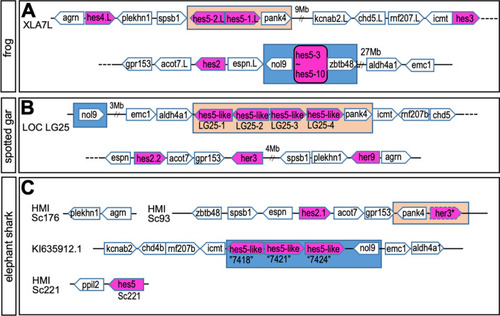
Syntenic analysis of |
|
Phylogenetic analysis of teleost |
|
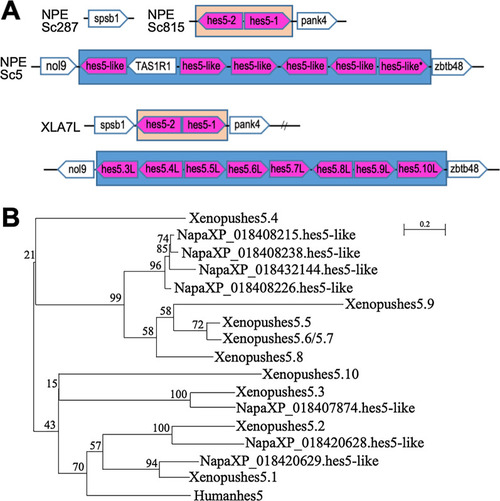
Comparison with |
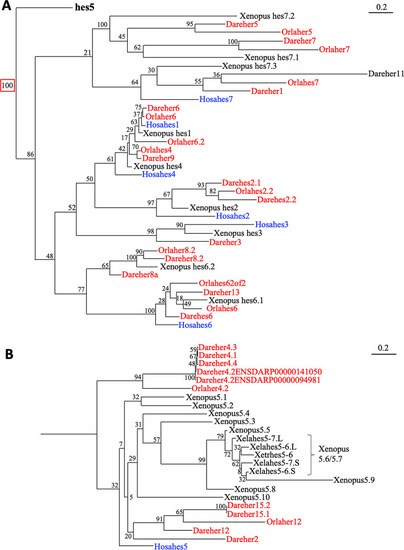
|
Phylogenetic analysis of |
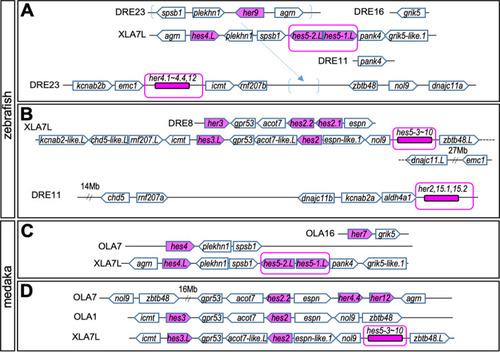
|
Comparison with |
|
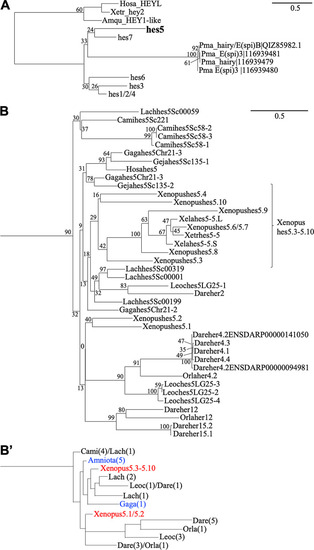
Comprehensive phylogenetic analysis of |
|
Syntenic and phylogenetic analysis of Tibetan frog |
|
Evolutionary acquisition of |