- Title
-
Functional in vivo characterization of sox10 enhancers in neural crest and melanoma development
- Authors
- Cunningham, R.L., Kramer, E.T., DeGeorgia, S.K., Godoy, P.M., Zarov, A.P., Seneviratne, S., Grigura, V., Kaufman, C.K.
- Source
- Full text @ Commun Biol
|
PHENOTYPE:
|
|
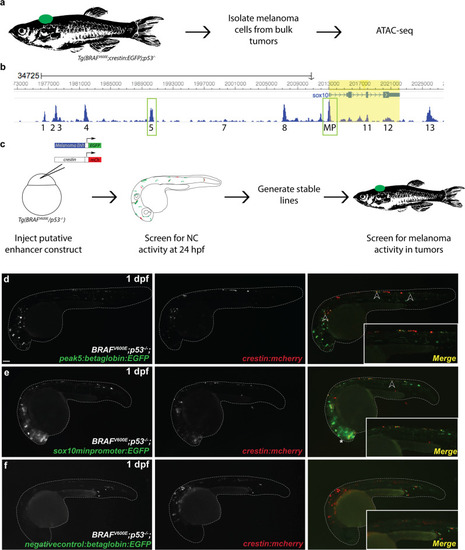
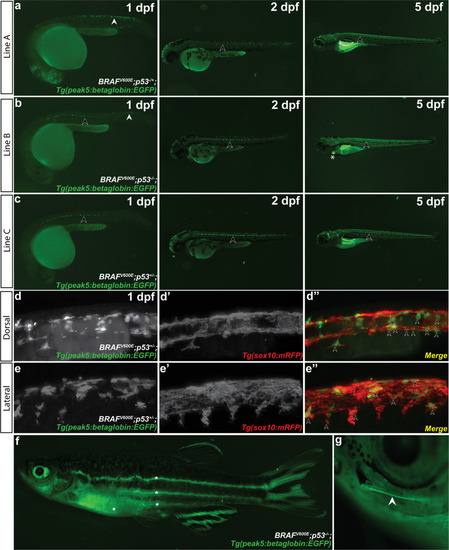
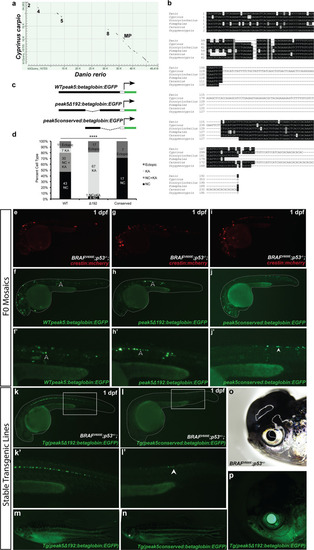
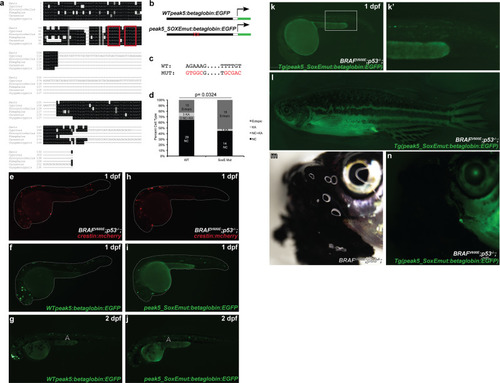
Multiple transgenic lines derived from independent founder (F0) parental fish were established that transmit the |
|
|
|
EXPRESSION / LABELING:
|
|
|
|
|