- Title
-
Hindbrain and Spinal Cord Contributions to the Cutaneous Sensory Innervation of the Larval Zebrafish Pectoral Fin
- Authors
- Henderson, K.W., Roche, A., Menelaou, E., Hale, M.E.
- Source
- Full text @ Front. Neuroanat.
|
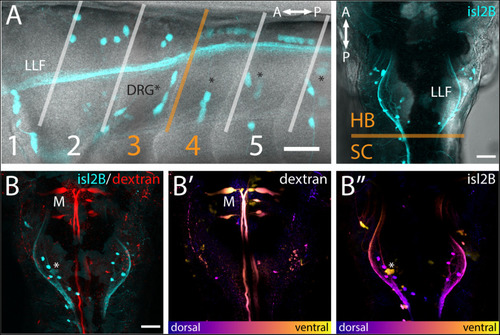
islet2B + neurons innervate the pectoral fins of 5 dpf larval zebrafish. |
|
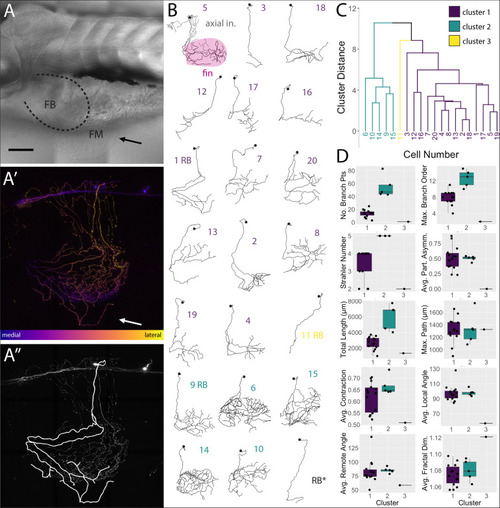
FSNs have somas in the hindbrain and spinal cord. |
|
FSNs exhibit a variety of soma morphologies. |
|
|
|
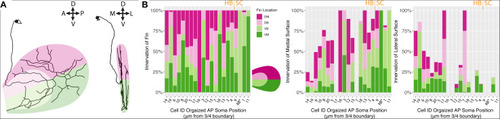
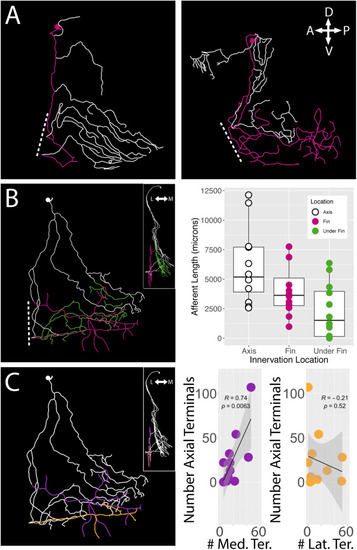
Fin sensory neurons exhibit biases to specific fin areas depending on soma location. |
|
|