- Title
-
The slc4a2b gene is required for hair cell development in zebrafish
- Authors
- Qian, F., Wang, X., Yin, Z., Xie, G., Yuan, H., Liu, D., Chai, R.
- Source
- Full text @ Aging (Albany NY)
|
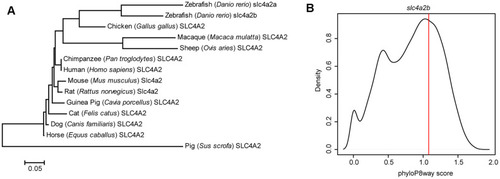
Evolutionary feature analysis of the slc4a2b gene. (A) Evolutionary relationships of SLC4A2 in different species. The evolutionary history was inferred using the Neighbor-Joining method, and the optimal tree with the sum of branch length = 1.66619414 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The analysis involved 14 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 237 positions in the final dataset. The evolutionary analysis was conducted in MEGA6. (B) slc4a2b phyloP8way conservation score (vertical red line) and the zebrafish protein coding gene phyloP8way conservation score density distribution (black line). The x-axis is the average phyloP8way score for the coding sequence region of the gene, and the y-axis is the density of the number of genes in each conservation score range. |
|
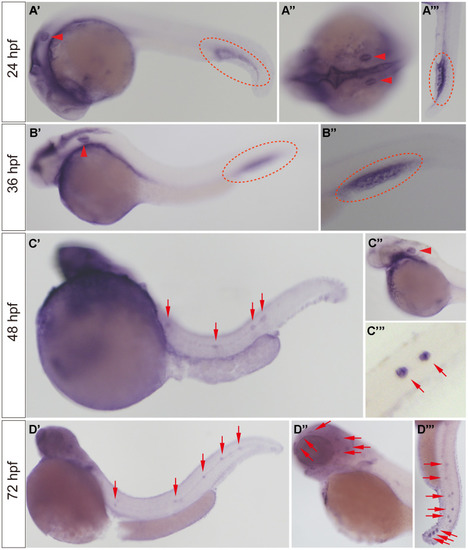
slc4a2b mRNA expression pattern detected by WISH. (A, B) The slc4a2b mRNA was mainly expressed in the otic vesicle (indicated by red arrowheads) and caudal vein (indicated by red dotted lines) at 24 hpf and 36 hpf. (A’’) shows the dorsal view of the otic vesicle, and (A’’’) and (B’’) are focused on the caudal vein. (C) At 48 hpf, slc4a2b mRNA was detected not only in the otic vesicle (indicated by the red arrowhead), but also in the lateral line neuromasts (indicated by red arrows). (C’’) and (C’’’) show the otic vesicle and the neuromasts at higher magnification, respectively. (D) slc4a2b mRNA was expressed in neuromasts at 72 hpf. (D’’) shows the neuromasts on the head (red arrowheads indicate the neuromast MI1, MI2, O2, et al.) and (D’’’) shows the posterior lateral line neuromasts. EXPRESSION / LABELING:
|
|
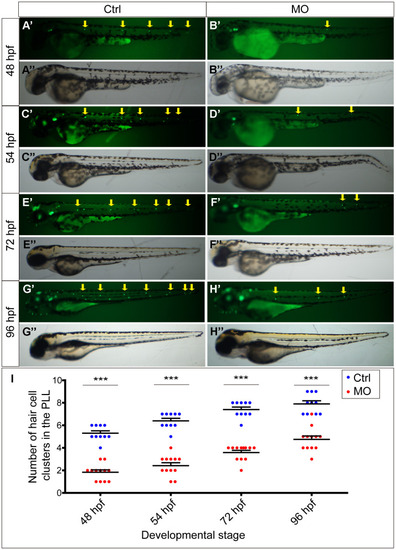
slc4a2b knockdown leads to decreased HC clusters in the posterior lateral line of zebrafish. (A–D) Zebrafish injected with slc4a2b-morpholino (MO) had normal morphology (B’’, D’’, F’’, H’’) but decreased HC clusters in the posterior lateral line (B’, D’, F’, H’) compared to the controls (Ctrl) (A’’, C’’, E’’, G’’ and A’, C’, E’, G’) at different developmental stages. The HC clusters in the posterior lateral line are indicated by yellow arrows. (I) Quantification of the number of HC clusters in the posterior lateral line in slc4a2b-morphants and controls at different developmental stages. ***P < 0.001. |
|
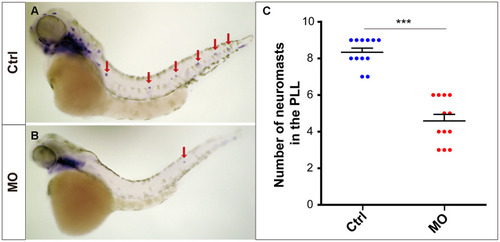
slc4a2b knockdown leads to decreased lateral line neuromasts in zebrafish. Zebrafish larvae at 72 hpf and the eya1 RNA probe were used in the WISH analysis. (A, B) slc4a2b-morphants (MO) (B) had fewer neuromasts (indicated by red arrows) compared to the controls (A). (C) Quantification of the number of the posterior lateral line neuromasts in slc4a2b-morphants and controls. ***P < 0.001. EXPRESSION / LABELING:
PHENOTYPE:
|
|
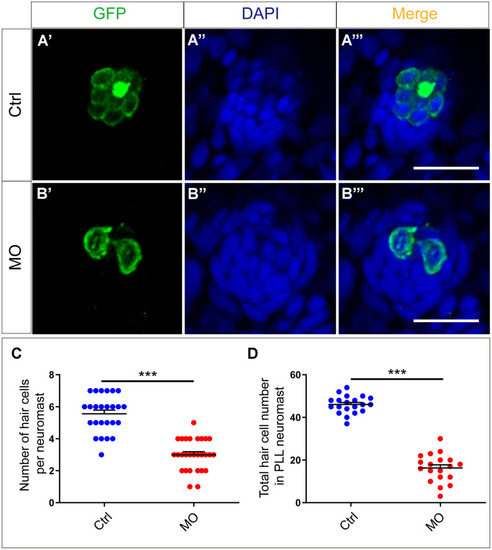
slc4a2b knockdown leads to decreased HCs in the posterior lateral line neuromast of zebrafish. (A, B) slc4a2b-morphants (MO) (B) had fewer HCs in each neuromast compared to controls (A). Tg(Brn3c:mGFP) transgenic zebrafish were used in the analysis, and the cells labeled by GFP represent HCs, while DAPI was used to stain the cell nuclei. (C, D) Quantification of the number of HCs in each neuromast and the total HCs in posterior lateral line neuromasts of the slc4a2b-morphants and controls. Scale bar = 20 μm, *** P < 0.001). EXPRESSION / LABELING:
PHENOTYPE:
|
|
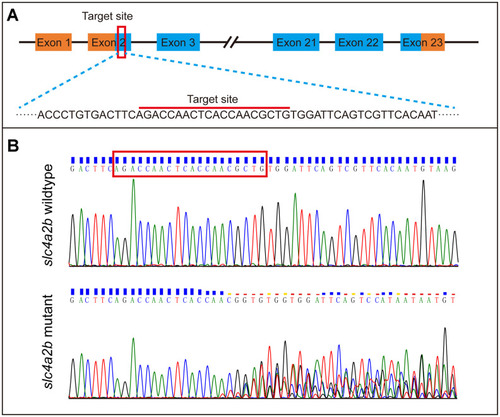
Generation of slc4a2b gene mutant zebrafish using CRISPR/Cas9 gene-editing technology. (A) The coding sequence in exon 2 of the slc4a2b gene was chosen to be the target of mutation. (B) Various mutations occurred in the target site of the slc4a2b gene in mutant zebrafish compared to the wild-type fish. |
|
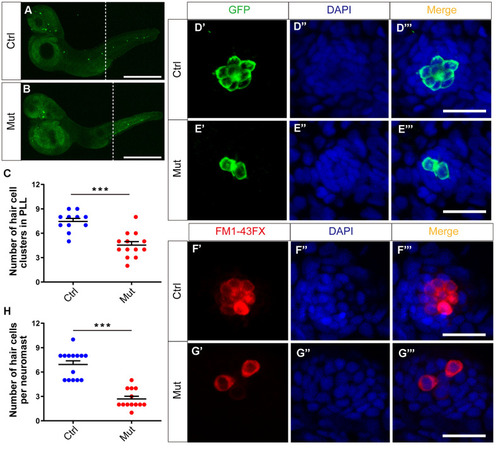
slc4a2b gene mutant zebrafish have fewer HC clusters and HCs in the posterior lateral line (PLL). (A, B) The Tg(Brn3c:mGFP);slc4a2bmut zebrafish had fewer HC clusters in the posterior lateral line compared to the controls. Here, the anti-GFP antibody was used to label the HCs. Scale bar = 500 μm. (C) Quantification of the number of HC clusters in the posterior lateral line of the slc4a2b gene mutant zebrafish and controls. ***P < 0.001. (D, E) The Tg(Brn3c:mGFP);slc4a2bmut zebrafish had fewer HCs in each posterior lateral line neuromast compared to controls. Here, anti-GFP antibody and DAPI were used to label the HCs and the nuclei, respectively. Scale bar = 20 μm. (F, G) The Tg(Brn3c:mGFP);slc4a2bmut zebrafish had fewer functional HCs in each posterior lateral line neuromast compared to controls. The successful staining with FM1-43FX dye was as a marker of functional HCs. Scale bar = 20 μm. (H) Quantification of the number of HCs in each neuromast of the slc4a2b mutant zebrafish and controls. *** P < 0.001. EXPRESSION / LABELING:
PHENOTYPE:
|
|
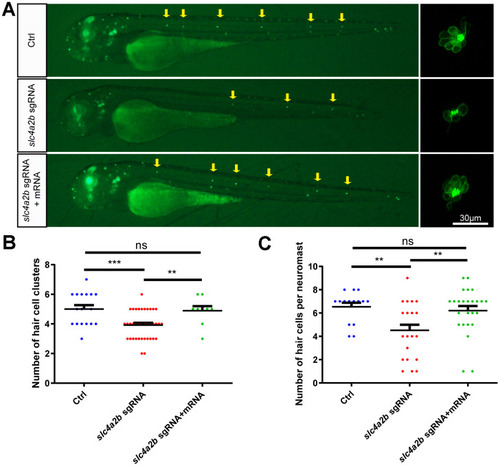
slc4a2b mRNA injection can rescue the phenotype induced by slc4a2b gene deficiency. The Tg(Brn3c:mGFP);slc4a2bmut zebrafish that were injected with Cas9 mRNA and slc4a2b sgRNA had fewer HC clusters (A, B) and fewer HCs in each posterior lateral line neuromast (A, C) compared to controls. However, slc4a2b mRNA injection could rescue the phenotype caused by slc4a2b mutation. *** P < 0.001; ** P < 0.01; ns, no significance. EXPRESSION / LABELING:
PHENOTYPE:
|
|
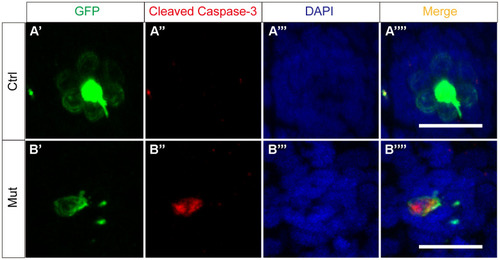
slc4a2b gene mutation leads to HC apoptosis. The control zebrafish, Tg(Brn3c:mGFP) (A), and the mutant zebrafish, Tg(Brn3c:mGFP);slc4a2bmut (B), were used for the apoptosis assay. Here, the anti-cleaved caspase-3 antibody was used to detect the apoptotic cells. Scale bar = 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|