- Title
-
Modeling of Wnt-mediated tissue patterning in vertebrate embryogenesis
- Authors
- Rosenbauer, J., Zhang, C., Mattes, B., Reinartz, I., Wedgwood, K., Schindler, S., Sinner, C., Scholpp, S., Schug, A.
- Source
- Full text @ PLoS Comput. Biol.
|
|
|
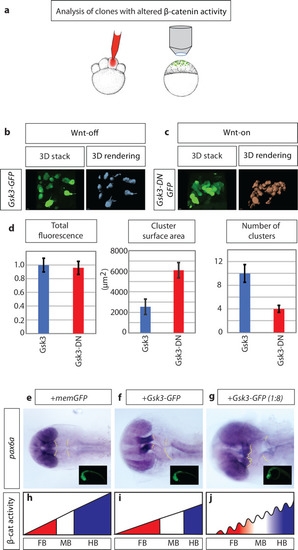
a) schematic illustration of the experimental procedure: 150ng mRNA injection of β-catenin effector Gsk3β into one cell at the eight blastomere stage. At sphere stage (prior induction of Wnt ligand expression), embryos were subjected to an image-based approach to analyse the distribution of cell clones in the animal tissue. b) expression of Gsk3β-GFP (Wnt-OFF) leads to a dispersed clone at the sphere stage. c) However, expression of dominant-negative Gsk3β-GFP (Gsk3β-DN-GFP; Wnt-ON) leads to clustering of the clonal cells. d) Gsk3β-DN expressing cells (Wnt-ON) show large clusters: demonstrated by an increased cluster surface and a reduced number of total clusters/cells compare to cells expressing WT Gsk3β (Wnt-OFF). The expression levels of Gsk3β and Gsk3β-DN are kept at a similar level shown by comparable total GFP-fluorescence in the clones. e)-g), embryos were injected with the indicated constructs and subjected to |
|
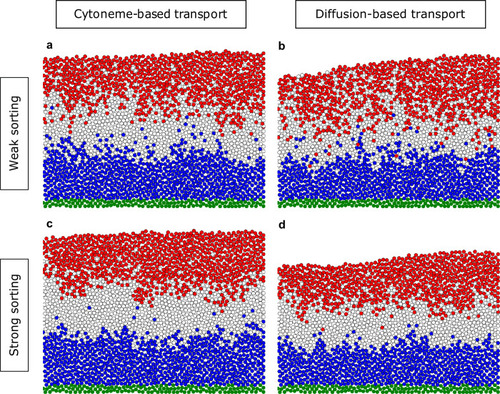
Distributions of the final cell fate at t = tTRS = 180 min in the tissue for different parameters of the cell dynamics and different transport mechanisms. Directed migration with the sorting parameter |
|
To determine the number of apoptotic cells in the developing embryo, immunohistochemical staining against Caspase3 is performed at 5 hpf, 7 hpf, and 9 hpf. |
|
In |
|
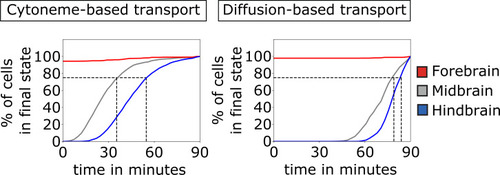
Comparison of the establishment of the three brain primordia based on cytoneme and diffusion-based transport. The percentage of cells that reach sufficient morphogen concentration to adopt their final cell fate (which is in this case determined by thresholding at tTRS = 90 min so that the tissue is split into thirds by cell numbers) is plotted over time. This is shown for forebrain (red solid line), midbrain (gray solid line), and hindbrain (blue solid line). The gray dashed lines mark the points in time where 75% of the midbrain and hindbrain cells adopted their final fate, respectively. After about 45 min, the majority of cells (75%) acquire their final fate if the ligand is transported on cytonemes. A diffusion-based distribution requires considerably longer time (about 80 min) to determine the cellular fate in the target tissue. The values are averaged over 100 simulations. |
|
To determine the temporal function Wnt/β-signaling on patterning, zebrafish embryos are treated with 5 μM of the Porcupine inhibitor IWP12 to block Wnt secretion. |
|
The field in which the tissue is modeled consists of precomputed non-overlapping potential cell positions (“fixed irregular lattice sites”). To initialize a simulation one margin of the field is filled with non-dividing morphogen producing cells (here shown in green), as well as morphogen receiving cells (shown in blue) with no initial morphogen concentration. After initialization (up to) four processes are sequentially executed for one timestep. In the |