- Title
-
Enhanced in vivo-imaging in medaka by optimized anaesthesia, fluorescent protein selection and removal of pigmentation
- Authors
- Lischik, C.Q., Adelmann, L., Wittbrodt, J.
- Source
- Full text @ PLoS One
|
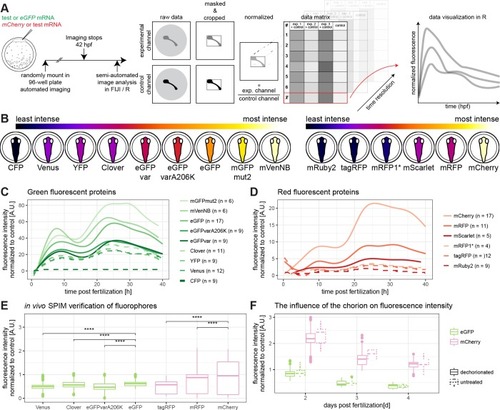
(A) Embryos were injected with a green fluorescent protein mRNA to be tested and mCherry mRNA or with eGFP mRNA and a red fluorescent protein mRNA to be tested (1-cell stage modified from [ |
|
(A) Relative comparison of |
|
(A) Schematic overview of the mean of the tested fluorescent proteins. The lookup table is indicated at the top of the panel, the green fluorescent proteins are shown on the left hand side and the red fluorescent proteins are shown on the right hand side (embryos modified from [ |
|
(A) Embryos were injected with |
|
(A) Medaka zygotes were microinjected with Cas9 mRNA and an injection mix according to |
|
Using the pigment double knockout in combination with α-Bungarotoxin, eGFP and mCherry coupled to histone2a the imaging was conducted in a single plane illumination microscope (SPIM) we could image the head region of wild-type and |