- Title
-
Glucocorticoid Treatment Leads to Aberrant Ion and Macromolecular Transport in Regenerating Zebrafish Fins
- Authors
- Schmidt, J.R., Geurtzen, K., von Bergen, M., Schubert, K., Knopf, F.
- Source
- Full text @ Front Endocrinol (Lausanne)
|
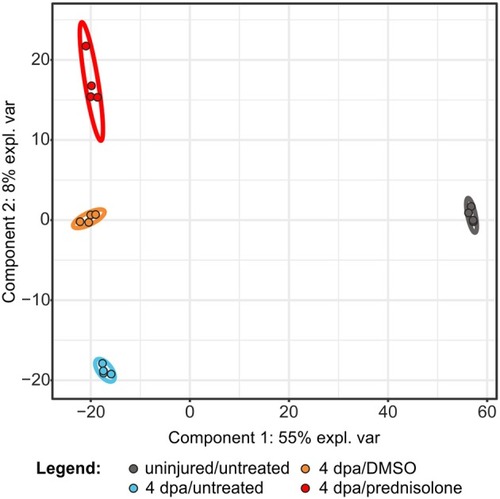
Partial least square discriminant analysis using quantitative values of all identified proteins. expl. var., explained variance. |
|
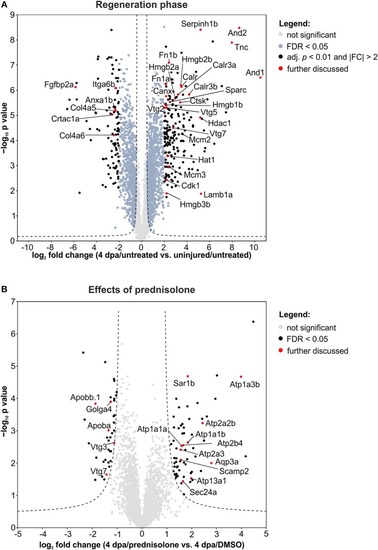
Volcano plots of all quantified proteins for the given analyses with their log2-transformed ratios of the mean centered abundances (fold changes, FC) and –log10-transformed |
|
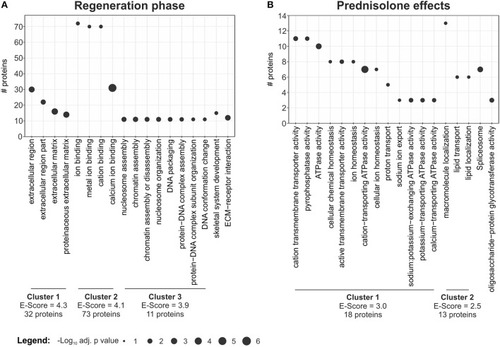
Term enrichment and clustering. Extracted enriched terms and clusters for the given analyses. Significance for enrichment (-log10-transformed adj. |
|
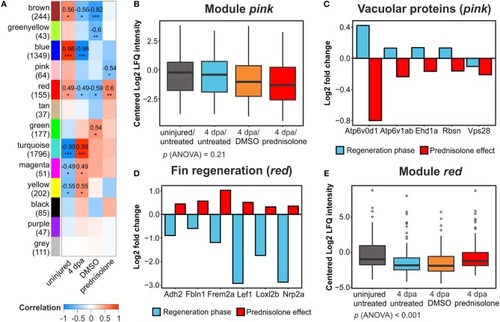
Weighted gene correlation network analysis of all filtered proteins. |
|
|
|
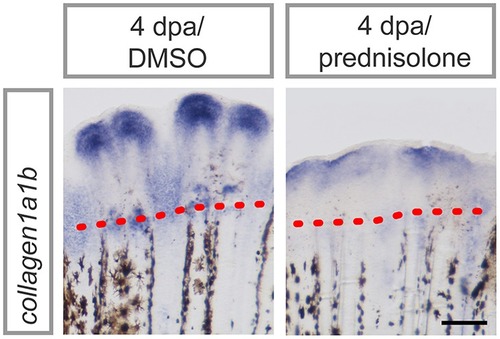
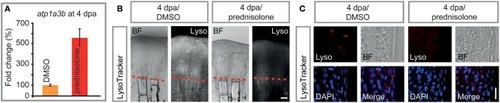
Impaired ATPase expression and lysosomal acidification. |