- Title
-
MIP/Aquaporin 0 Represents a Direct Transcriptional Target of PITX3 in the Developing Lens
- Authors
- Sorokina, E.A., Muheisen, S., Mlodik, N., and Semina, E.V.
- Source
- Full text @ PLoS One
|
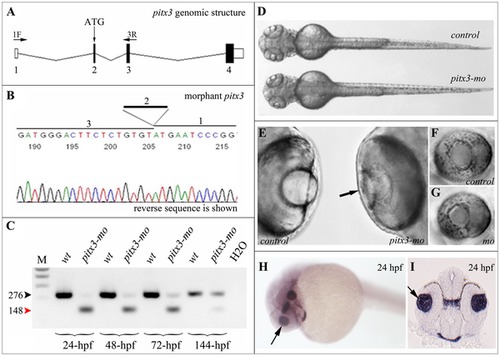
Injection of pitx3 morpholino results in abnormal splicing of pitx3 transcript and small lens phenotype. A. Schematic drawing of pitx3 gene; position of initiation codon (ATG) and RT-PCR primers (1F and 3R) are indicated, exons are numbered. B. Sequencing of the pitx3-mo transcript generated with primers located in the first and third pitx3 exons demonstrates absence of exon 2 in the resultant product. C. RT-PCR results obtained with pitx3 1F/3R primers using RNA extracted from embryos injected with control or pitx3 morpholino. Please note a strong decrease in normal 276-bp product (black arrowhead) and presence of abnormal 148-bp product (red arrowhead) in pitx3-mo samples; hpf- hours post fertilization of analyzed embryos. D–G. Morphological phenotypes of zebrafish pitx3-mo embryos. In comparison to control embryos, a smaller head can be observed in pitx3 morphants (72-hpf embryo is shown; D) as well as an obvious reduction in lens size (black arrow) at later stages (96-hpf embryos are shown; E–G). H and I. Expression of pitx3 in the developing lens in 24-hpf embryos. Please note robust expression in the lens vesicle (arrows) as demonstrated by both whole mount (H) and section (I) in situ. EXPRESSION / LABELING:
PHENOTYPE:
|
|
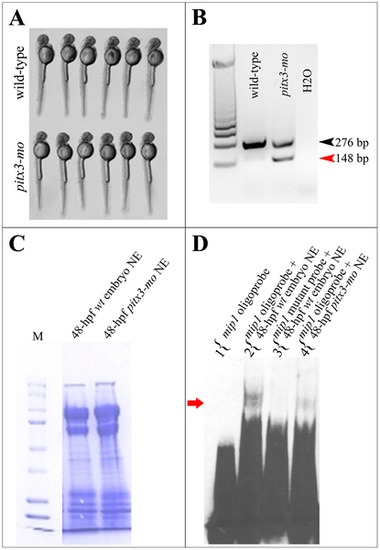
Formation of high molecular weight mip1 promoter- protein complexes is dependent on pitx3 presence. A. 48-hpf wild-type and pitx3 morphant embryos showing normal body length and morphology that were selected for EMSA experiments. B. Results of RT-PCR performed with RNA extracted from the pooled tail tissues from wild-type and pitx3 morphant embryos shown in C. A sharp reduction in normal pitx3 transcript (black arrowhead) and the presence of abnormally spliced product (red arrowhead) are evident in pitx3 morphant embryos. C. Coomassie Blue R-250 stained polyacrilamide gel demonstrating equal protein concentration in nuclear extracts obtained from wild-type (lane 1) and pitx3 morphant (lane 2) embryos that were used in EMSA experiments shown in A. D. Electrophoretic mobility shift assays (EMSA) show formation of a DNA-protein complex when an oligonucleotide corresponding to the -44/-76 region of zebrafish mip1 promoter and nuclear extracts from 48-hpf wild-type zebrafish embryos are used. Please note a presence of a specific slow migrating complex, which is formed by wild-type mip1 probe and proteins extracted from nuclei of 48-hpf wild-type zebrafish embryos (lane 2), absence of this complex in lane 3 when the same nuclear extracts were combined with a mutant mip1 probe where the pitx3-binding bicoid site GGATTA was replaced by AAATTA, and sharp reduction of this complex in lane 4 containing a combination of a wild-type mip1 probe and nuclear extracts obtained from pitx3 morphants (red arrow). |
|
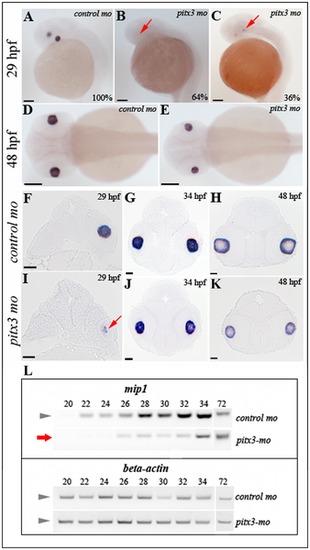
Analysis of mip1 expression in pitx3-mo and control embryos via in situ hybridization and RT-PCR. A, D, F-H. Normal mip1 expression in control-injected embryos at 29-, 34- and 48-hpf. B, C, E, I–K. Altered mip1 expression is observed in pitx3 morphants at 29-hpf with 64% of embryos demonstrating a complete absence of mip1 expression (B) and the remaining larvae showing markedly reduced mip1 expression (C and I). Reduced mip1 expression is also observed in 34- and 48-hpf embryos (E, J, K). Red arrows show sites of expected mip1 expression. Scale bars: A–E: 100 μM; F–L: 20 μM. L. Results of semi-quantitative RT-PCR showing reduced expression of mip1 in pitx3 morphants at early stages of development (red arrow). EXPRESSION / LABELING:
|

Unillustrated author statements PHENOTYPE:
|