- Title
-
Identification and functional analysis of the vision-specific BBS3 (ARL6) long isoform
- Authors
- Pretorius, P.R., Baye, L.M., Nishimura, D.Y., Searby, C.C., Bugge, K., Yang, B., Mullins, R.F., Stone, E.M., Sheffield, V.C., and Slusarski, D.C.
- Source
- Full text @ PLoS Genet.
|
Identification of a second BBS3 transcript. (A) Alignment of human BBS3 and BBS3L proteins. (B) C-terminal end alignment of human, mouse, and zebrafish BBS3L protein. Asterisks (*) indicate identical amino acids shared in all alignments, while colons (:) and periods (.) represent conserved amino acids. (C) RT–PCR tissue expression profile of zebrafish bbs3 and bbs3L transcripts in wild-type adult zebrafish tissues: fat, brain, heart, whole eye and retina. β-actin was used as a positive control. bbs3 is expressed in all adult tissues examined, while bbs3L expression is limited to the eye. (D) RT–PCR developmental expression profile of bbs3 and bbs3L at the following stages: maternal, 8–12 somites, 24, 36, 42, 48, 60, 72, 96 hpf, and 5 dpf. bbs3 is expressed throughout development, while the long form is only present by 48 hpf, correlating with photoreceptor development. |
|
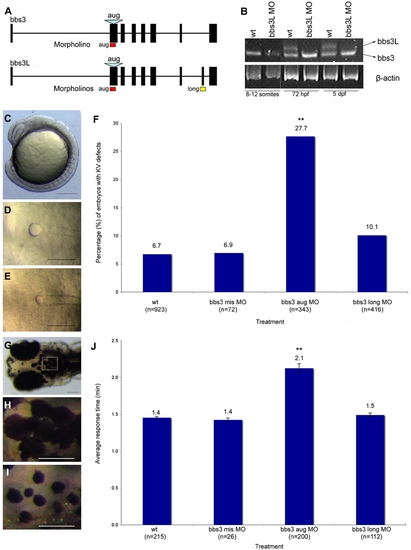
bbs3 gene structure and cardinal features of BBS knockdown in zebrafish. (A) Schematic depicting the bbs3 gene structure and antisense oligonucleotide strategy used to target either both transcripts (bbs3 aug MO) or to target only bbs3L (bbs3 long MO) in zebrafish embryos. The bbs3 aug MO targets the start site of the gene and thus hits both transcripts, while the bbs3L MO is a splice-blocking morpholino that only targets the long form. (B) RT-PCR from staged bbs3L morphant embryos at 8–12 somites, 72 hpf, and 5 dpf. The bbs3L transcript is absent through 5 dpf injected embryos indicating successful knockdown. Note that the bbs3 transcript is unperturbed in bbs3L morphants. (C–E) Images of live zebrafish embryos at 8–10 somite stage. Scale bar 200 μm. (C) Side view of an embryo highlighting the location of the Kupffer′s Vesicle (circle), the ciliated structure located in the tailbud. (D) Dorsal view of a normal sized KV from a wild-type embryo. (E) bbs3 aug MO–injected embryos with a reduced KV. (F) The percentage of embryos with KV defects (reduced or absent) in uninjected, control MO, bbs3 aug MO and bbs3 long MO injected embryos. The sample size (n) is noted on the x-axis. **Fisher′s Exact test, p<0.001. (G–I) Epinephrine-induced melanosome transport of wild-type 6-day old larvae. Scale bar 100 μm. (G) Melanosome transport is observed in cells on the head of the embryos. Boxed region is magnified for (H,I). (H) Wild-type larvae prior to epinephrine treatment and (I) the endpoint at 1.4 minutes after epinephrine treatment. (J) Epinephrine-induced retrograde transport times. The sample size (n) is noted on the x-axis. **ANOVA with Tukey, p<0.01. PHENOTYPE:
|
|
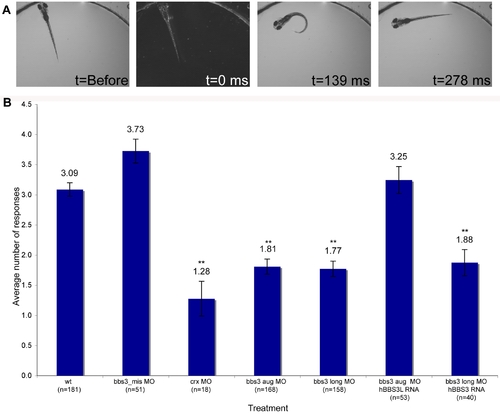
Vision startle response in zebrafish. (A) Vision function was assayed in 5-day old embryos by testing embryos sensitivity to short blocks in light at 30 second intervals for 5 trials (adapted from Easter and Nicola 1996). Selected images from a time-lapse collection before and immediately after a one second block in light. The typical response, a distinct C-bend, is scored as a positive response as shown in time point 139 ms. ms, milliseconds. (B) Quantification of the vision startle response for each treatment. Cone-rod homeobox (crx) gene knockdown was used as a control for vision impairment. bbs3 morphants lacking either both transcripts or only the long transcript showed a statistically significant reduction in the number of responses, indicating visual impairment. Rescue experiments using wild-type human BBS3L or BBS3 RNA co-injected with the bbs3 morpholinos demonstrated that hBBS3L RNA is sufficient to rescue the vision defect associated with knockdown, while hBBS3 is not sufficient to rescue the vision defect. The sample size (n) is noted on the x-axis. **ANOVA with Tukey, p<0.01. PHENOTYPE:
|
|
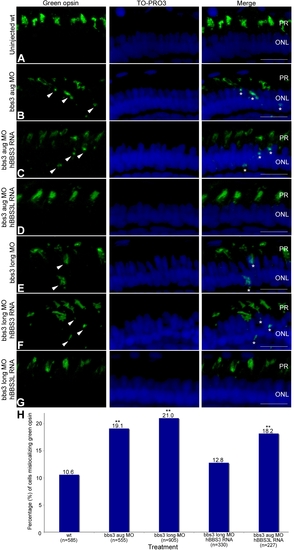
Green opsin mislocalization and rescue in 5-day-old bbs3 morphant zebrafish. Immunofluorescence of green cone opsin (green) on transverse cryosections of 5-day-old embryos (A) uninjected wild-type, (B) bbs3 aug MO, (C) bbs3 aug MO and hBBS3 RNA, (D) bbs3 aug MO and hBBS3L RNA, (E) bbs3 long MO, (F) bbs3 long MO and hBBS3 RNA, and (G) bbs3 long MO and hBBS3L RNA. To-Pro3 was used to counterstain the nuclei (blue). In bbs3 aug (B) and bbs3 long morphants (E), green opsin was not restricted to the outer segment of the photoreceptors and was detected in the cell bodies of the outer nuclear layer (arrowheads and asterisks). Expression of hBBS3L RNA improved green opsin localization in both bbs3 aug (D) and bbs3L (G) morphants. Of note, hBBS3 RNA failed to rescue green opsin localization in bbs3 aug (C) and bbs3 long (F) morphant embryos (arrowheads and asterisks). OS, outer segment; ONL, outer nuclear layer. Scale Bar 10 µm. (H) The percentage of mislocalizing green opsin cells. The sample size (n) is noted on the x-axis and represents the total number of green opsin positive cells counted. **Fisher′s exact test, p<0.01. EXPRESSION / LABELING:
|