- Title
-
5' modifications improve potency and efficacy of DNA donors for precision genome editing
- Authors
- Ghanta, K.S., Chen, Z., Mir, A., Dokshin, G.A., Krishnamurthy, P.M., Yoon, Y., Gallant, J., Xu, P., Zhang, X.O., Ozturk, A.R., Shin, M., Idrizi, F., Liu, P., Gneid, H., Edraki, A., Lawson, N.D., Rivera-Pérez, J.A., Sontheimer, E., Watts, J.K., Mello, C.C.
- Source
- Full text @ Elife
|
(A) Schematic showing the TLR assay to quantify HDR efficiencies using unmodified or end-modified double-stranded DNA (dsDNA) donors. Editing efficiencies plotted as percentage of (B) green fluorescent protein (GFP)+ (HDR) and (C) mCherry+ (non-homologous end joining [NHEJ]) HEK293T TLR cells at different amounts of unmodified, 2?-O-methyl (2?OMe)-RNA::triethylene glycol (TEG)-modified dsDNA donors. Editing efficiencies plotted as percentage of (D) GFP+ (HDR) and (E) mCherry+ (NHEJ) HEK293T TLR cells at 1.2 pmol of dsDNA donors indicated. Percentage of GFP+ cells obtained with dsDNA donors modified with various lengths of ethylene glycol (F) and with modifications to only one end or both 5?-ends of the donor. TS, target strand; NTS-,non-target strand (G). Mean ± s.d. for at least three independent replicates are plotted; two replicates for TEG donor in panel G.
|
|
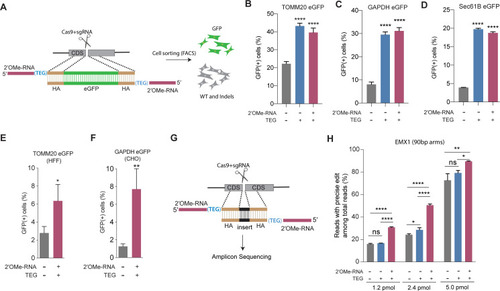
(A) Schematic representation of the 5?-modified donor design for enhanced green fluorescent protein (eGFP) insertion and fluorescence-activated cell sorting (FACS) is shown. Efficacy of eGFP integration at (B) TOMM20 and (C) GAPDH (D) Sec61B loci in HEK293T cells using unmodified, triethylene glycol (TEG) or 2?-O-methyl (2?OMe)-RNA::triethylene glycol (TEG)-modified donors are plotted as percentage of GFP+ cells. Efficacy of eGFP integration at the (E) TOMM20 locus in human foreskin fibroblast (HFF) (747 bp knock-in with ~1 kb homology arms) and (F) Gapdh locus in Chinese hamster ovary (CHO) (1635 bp knock-in with ~800 bp homology arms) cells using double-stranded DNA (dsDNA) (500 ng) donors with and without 2?OMe-RNA::TEG modifications at the 5?-ends. (G) Schematic representation of the dsDNA donor design used for quantification with deep sequencing is shown. (H) Illumina sequencing reads with precise knock-in are plotted for dsDNA donors with 90 bp homology arms at EMX1 locus in HEK293T cells. Mean ± s.d. for at least three independent replicates are plotted. p-Values were calculated using one-way ANOVA and in all cases end-modified donors were compared to unmodified donor unless indicated otherwise (Tukey?s multiple comparisons test; ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; ns, not significant).
|
|
(A) Editing efficacy plotted as percentage of green fluorescent protein (GFP+) (precise) HEK293T traffic light reporter (TLR) cells at different amounts of unmodified and 2?-O-methyl (2?OMe)-RNA::triethylene glycol (TEG)-modified long single-stranded DNA (ssDNA) donors (800 nt). (B) Editing efficacy of GFP-to-blue fluorescent protein (BFP) reporter conversion in K562 cells using different amounts of unmodified and 2?OMe-RNA::TEG-modified 66 nt single-stranded oligodeoxynucleotide (ssODN) donors plotted as percentage of BFP+ cells (homology-directed repair [HDR]). (C). Editing efficacy of GFP-to-BFP conversion in K562 cells using 0.5 pmol of ssODN donors modified at the 5?-end alone, the 3?-end alone, or at both the 5?- and 3?-ends, with phosphorothioate (PS), TEG, 2?OMe-RNA, or 2?OMe-RNA::TEG, plotted as percentage of BFP+ cells (precise). Complete figure of panel C is shown, along with other modifications, in Figure 3?figure supplement 2. Mean ± s.d. for at least three independent replicates are plotted. p-Values were calculated using one-way ANOVA and in all cases end-modified donors were compared to unmodified donor unless indicated otherwise (Tukey?s multiple comparisons test; ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; ns, not significant).
|
|
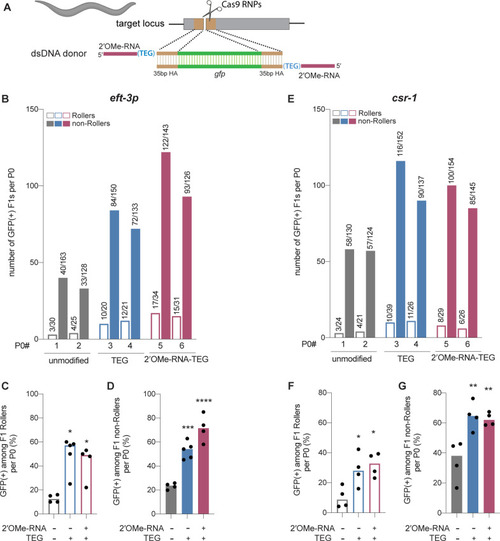
(A) Schematic showing end-modified double-stranded DNA (dsDNA) donors (25 ng/µl) with short (~35 bp) homology arms to insert gfp tag. (B) Number of green fluorescent protein (GFP) expressing animals among entire F1 brood of two representative P0 animals for each donor type are plotted for eft-3p reporter locus. Fraction of F1 animals expressing GFP among (C) Roller and (D) non-Roller cohorts are plotted as percentage for eft-3p locus. Similarly, (E) number of GFP expressing animals among two representative broods, fraction of F1 animals expressing GFP among (F) Roller and (G) non-Roller cohorts are plotted for csr-1 locus. Open bars (Rollers) and closed bars represent (non-Rollers) median. Number of GFP expressing animals among total number of animals scored per cohort are shown above the bars. n ? 4 broods for each donor condition. p-Values were calculated using one-way ANOVA and in all cases end-modified donors were compared to unmodified donors (Tukey?s multiple comparisons test; ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; ns, not significant).
|
|
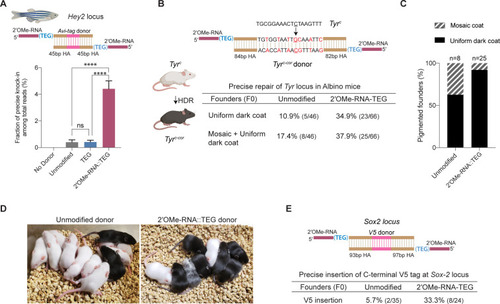
(A) Unmodified, TEG, and 2?-OMe-RNA::TEG-modified double-stranded DNA (dsDNA) donors were injected into zebrafish zygotes. dsDNA donor design to knock-in Avi-tag is shown on the top and the fraction of Illumina reads containing precise knock-in are plotted as percentages. Mean ± s.d. for at least three independent replicates (two for unmodified donors) are plotted (B). Design of the dsDNA donors injected into mouse zygotes to precisely convert the coat color of albino mice (TyrC) to pigmented (TyrC-Cor) by editing C to G (underscored) along with six silent mutations (in red) is shown. Percentages of F0 founder mice with black coat are shown. (C) Percentages of animals among homology-directed repair (HDR)-positive F0s that have uniform dark coat or mosaic coat color are plotted for unmodified and 5?-modified donors. (D) Representative pictures of 10-day- old F0 mice with pigmented (HDR) or white (wild-type [WT] or indel) coat color are shown. One mosaic mouse (third from left) can be seen among the pups obtained with end-modified donor. (E) Donor design to knock-in V5 tag at the C-terminus of Sox2 is shown on the top. Percentage of founder animals containing perfect V5 insertion at Sox2 locus are shown for each donor type. HA, homology arms. p-Values were calculated using one-way ANOVA (Tukey?s multiple comparisons test; ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; ns, not significant).
|
|
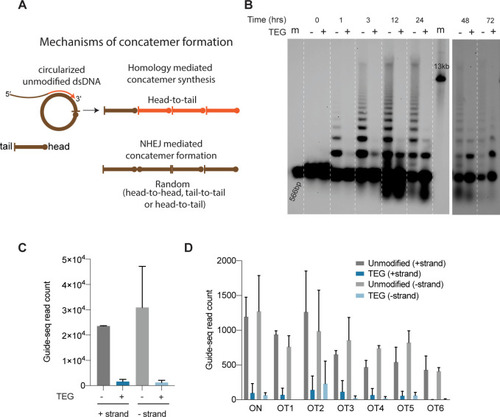
(A) Model for mechanisms of concatemer formation for unmodified donors is shown. (B) Southern blot of unmodified and triethylene glycol (TEG)-modified double-stranded DNA (dsDNA) (566 bp) nucleofected into HEK293T cells and collected at indicated time points. Concatemerization of unmodified DNA is visualized as ladders; 566 bp DNA and 13 kb long DNA are used as size markers (m). Number of GUIDE-seq reads with unmodified and TEG-modified short dsDNA (34 bp) integration for (C) whole genome and (D) on-target (ARHGEF9) and six previously validated off-target loci are plotted. Data from two biological replicates is shown.
|