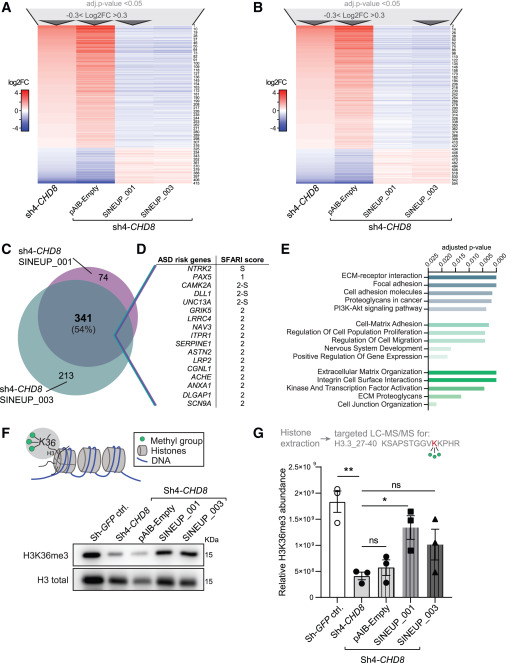
Fig. 2 SINEUP-CHD8 molecules rescue cellular and molecular phenotypes associated with CHD8 suppression (A and B) Heatmaps of deregulated genes upon CHD8 knockdown in Sh4-CHD8 and Sh4-CHD8 hNPCs cells transduced with lentiviral particles expressing the pAIB-Empty control vector, rescued through SINEUP_001-mediated (A) and SINEUP_003-mediated (B) CHD8 protein translation induction (n = 4 replicates per condition, all DEGs first filtered for adjusted p value < 0.05 and −0.3 < log2FC > 0.3 as indicated by dark gray arrows, resulting in a total of N = 415 and N = 554 responding DEGs, respectively. (C) Area proportional Venn diagram demonstrating overlap of number of DEGs and percent of overlap between rescued transcripts (at padj < 0.05 and −0.3 < log2FC > 0.3) in Sh4-CHD8 knockdown cells. (D) Selection of ASD-risk genes found among the N = 341 shared DEGs responding to SINEUP_001 and SINEUP_003 expression in Sh4-CHD8 hiNPCs (C) and their SFARI gene score. (E) Selected terms from KEGG 2021 (teal, top), GO-term biological process 2023 (aquamarine, middle), and Reactome 2022 (green, bottom) gene list annotation as performed by the Enrichr tool54,55,56 on the N = 341 shared DEGs represented in (C). (F) CHD8 regulates H3K36 tri-methylation levels of cells chromatin (scheme, top), WB visualization of H3K36me3 levels in acidic Histone extracts upon hiNPC transduction (bottom). (G) Scheme of targeted LC-MS/MS approach to quantify H3K36me3 levels in histone extracts upon hiNPC transduction. A parallel reaction monitoring (PRM) method was established to target the trimethylated form of KSAPSTGGVKKPHR (H3.3_27–40) peptide. Skyline (version 24.1) was used to analyze and normalize peptide abundance changes (top). Relative abundance of H3K36me3 levels as obtained by targeted LC-MS/MS experiments (bottom) (n = 3 replicates per condition, data are presented as scatter dot blot ± SEM; one-way ANOVA followed by Sidak’s test for multiple testing correction, significance level ∗∗p ≤ 0.01, ∗p ≤ 0.05; ns, not significant).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Mol. Ther.