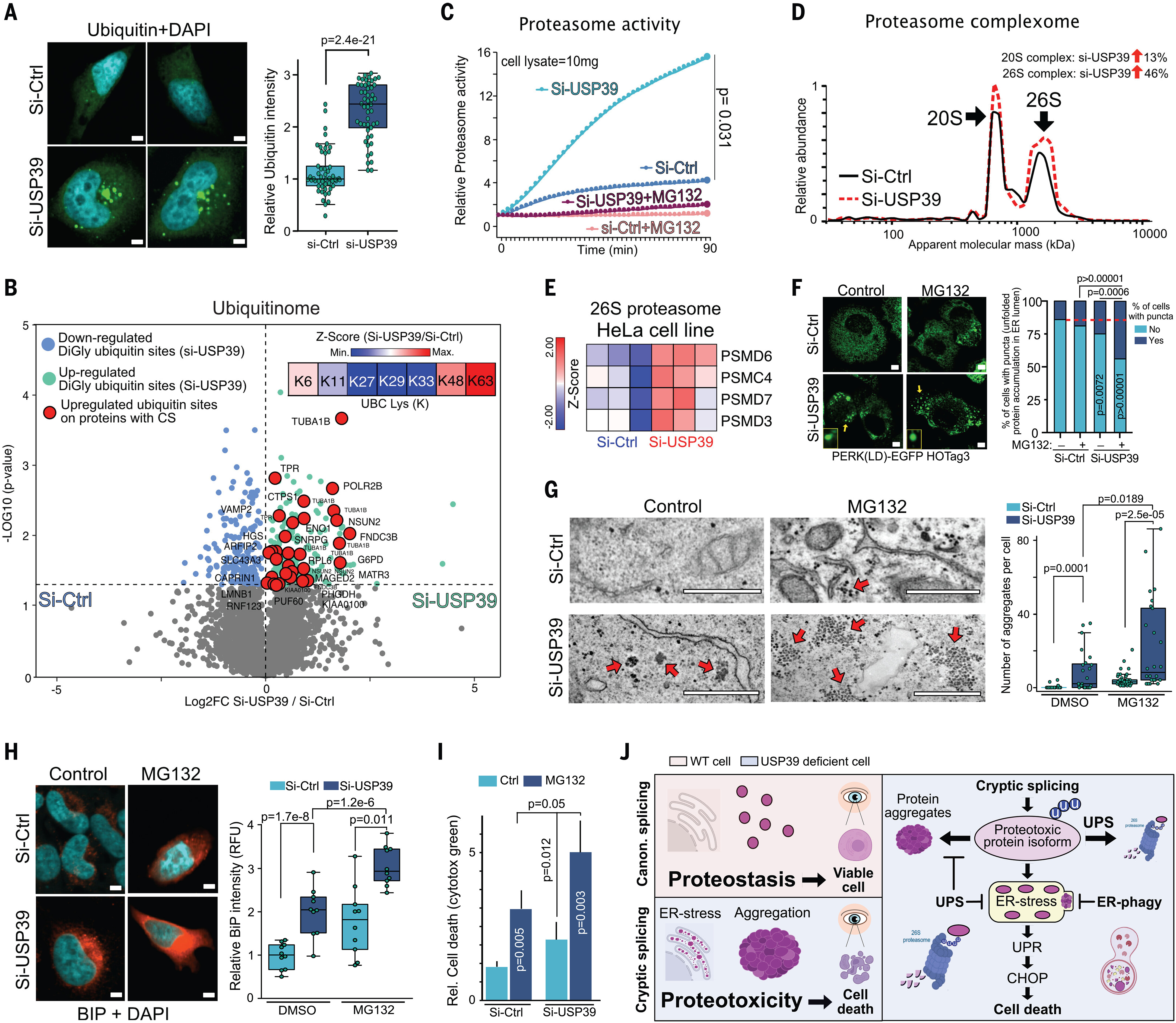
Fig. 5 The UPS degrades misspliced isoforms to mitigate proteotoxicity. (A) Ubiquitin (green) immunofluorescence (left) and quantification (right) in si-USP39 and si-Ctrl HeLa cells (n = 50; two-tailed t test; scale bars are 6 μm). (B) Volcano plots representing ubiquitinated diGly sites of si-USP39 and si-Ctrl cells (red indicates significantly more diGly ubiquitin sites in proteins translated from misspliced transcripts in si-USP39 cells). In the right corner, a z-score heat map of K6, K11, K27, K29, K33, K48, and K63 polyubiquitin chain abundances in si-USP39 cells is shown (n = 3). (C) Relative 7-amino-4-methylcoumarin (AMC) levels (proteasome activity) after the incubation of 10 mg of si-USP39 and si-Ctrl cell lysates with 50 μM AMC-Suc-LLVY probe. MG132 was added as a control (data are means ± SD; n = 3; two-tailed t test). (D) Relative protein abundance of 20S and 26S proteasome complexes in si-Ctrl and si-USP39 HeLa cells from fig. S11A. (E) Heat map representing gene expression (z-score) of representative 26S proteasome genes in si-USP39 and si-Ctrl HeLa cells (n = 3). (F) Immunofluorescence (left) and quantification (right) of puncta in si-Ctrl and si-USP39 PERK(LD)-EGFP-HOTag3 HeLa reporter cells exposed to MG132 (10 μM) for 10 hours (si-Ctrl n = 203, si-Ctrl+MG132 n = 188, si-USP39 n = 186, and si-USP39+MG132 n = 181; Fisher’s exact test calculator; yellow arrows mark the puncta of representative cells that have puncta; scale bars are 3 μm). The red dashed line indicates the percentage of cells with puncta for si-Ctrl cells. (G) TEM images (left) and quantification (right) of aggregates of si-Ctrl and si-USP39 HeLa cells exposed to MG132 (10 μM for 8 hours) si-Ctrl n = 31, si-USP39 n = 24, si-ctrl+MG132 n = 41, and si-USP39+MG132 n = 26; two-tailed t test; arrows indicate aggregates; scale bars are 500 nm) (H) BiP immunofluorescence (red) (left) and quantification (right) in si-Ctrl and si-USP39 MG132 HeLa cells exposed to BiP (10 μM for 8 hours) (n = 10; two-tailed t test; scale bars are 5 μm). DMSO, dimethyl sulfoxide. (I) Cell death assay using cytotox green dye in si-Ctrl and si-USP39 HeLa cells (data are means ± SD; n = 3; two-tailed t test) exposed to MG132 (10 μM) for 20 hours. (J) Schematic model summarizing our findings. CS triggers proteotoxicity but is mitigated by UPS and ER-phagy. For box-and-whisker plots, the center line represents the median, box limits are upper and lower quartiles, and whiskers are minimum and maximum values. [Panel (J) created in part with BioRender.com]
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Science