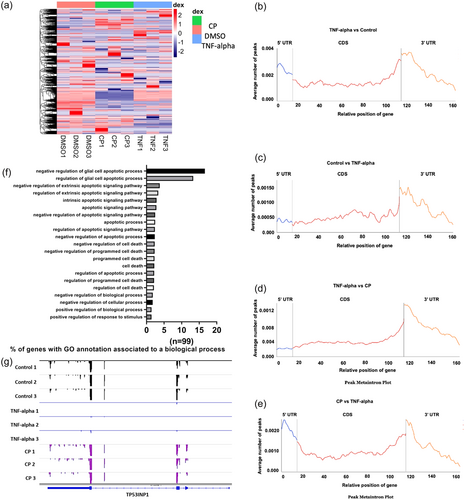
Fig. 2 m6A methylome analysis of TNF-α-treated HeLa cells. miCLIP-seq was used to obtain the m6A methylome of TNF-α treated HeLa cells as outlined in Materials and Methods. (a) Heat map of differentially m6A-methylated transcripts in CP- and TNF-α-treated HeLa cells. The list of differentially m6A-methylated transcripts in CP-treated cells was published previously (Alasar et al., 2022). Metaintron profiles of transcripts upregulated (b) or downregulated (c) in m6A methylation in TNF-α-treated HeLa cells and upregulated (d) or downregulated (e) in TNF-α-induced apoptosis compared to CP-treated HeLa cells along a normalized transcript, consisting of three rescaled nonoverlapping segments: 5′UTR, CDS, and 3′UTR. (f) Gene Ontology analyses of differentially m6A-methylated genes associated with apoptosis. (g) Distribution of m6A modification along the CDS, 3′UTR and 5′UTR of TP53INP1 mRNA was displayed by Integrative Genomics Viewer (IGV): Control (upper panels), as a control, TNF-α (middle panels) and CP treatment (lower panels).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ J. Cell. Physiol.