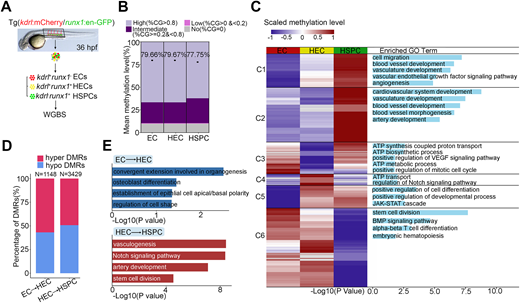
Fig. 1 Dynamic DNA methylation during HSPC generation. (A) Flowchart of sorting and whole genome bisulfite sequencing. EC, endothelial cell; HEC, hemogenic endothelial cell; HSPC, hematopoietic stem and progenitor cell; WGBS, whole genome bisulfite sequencing. (B) Global DNA methylation levels in EC, HEC, HSPC and fraction of CpGs displaying high (>0.8), intermediate (≥0.2 and <0.8), low (>0 and <0.2) and no (=0) 5mC. n=3 replicates. (C) Heatmap displays differentially methylated regions (DMRs) in EC, HEC and HSPC. Right column shows functional enrichment of DMR-related genes by GREAT analysis. The x-axis represents the negative log of the P-values of the enrichment of the corresponding GO terms. C1-C6, Clusters 1-6. (D) The percentage of hyper-DMRs and hypo-DMRs between each two consecutive stages: EC versus HEC and HEC versus HSPC. (E) GO enrichment of DMRs by GREAT analysis between each two consecutive stages: EC versus HEC and HEC versus HSPC. The x-axis represents the negative log of the P-values of the enrichment of the corresponding GO terms.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development