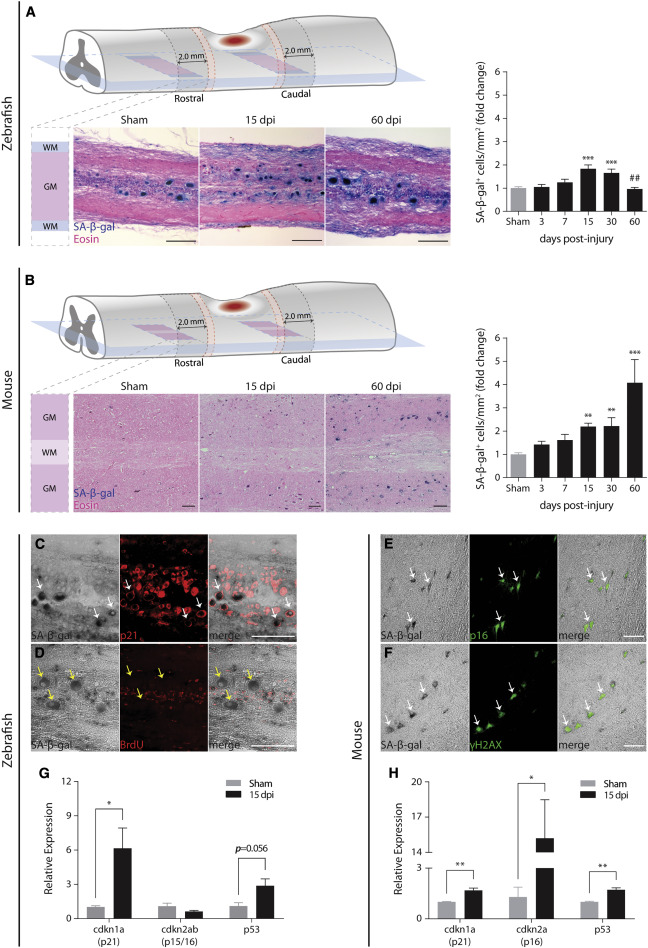
Fig. 1 Different senescent cell dynamics are induced after spinal cord injury in zebrafish and in mice (A and B) SA-β-gal+ cells (blue) were detected and quantified in non-injured (sham) and injured zebrafish spinal cords at different time points (3, 7, 15, 30, and 60 days post-injury [dpi]). n = 5–16. Similarly, SA-β-gal+ cells were detected and quantified in mouse laminectomized (sham) and injured spinal cords at the same time points. n = 4–16. Eosin counterstaining was performed after cryosectioning. Cells were quantified at the lesion periphery along 2.0 mm in longitudinal sections spanning the ventral horn. A 0.5-mm interval (red dashed zone) was established between the lesion border and the beginning of the quantification region. SA-β-gal+ cells were only quantified in the gray matter (GM) and not in the white matter (WM). Quantifications are presented as fold change toward sham. In zebrafish, SA-β-gal+ cells reach a peak at 15 dpi (238.6 cells/mm2), a 2-fold increase compared to sham (127.2 cells/mm2). In mice, SA-β-gal+ cells display a 4-fold increase at 60 dpi (80.0 cells/mm2) toward sham (19.0 cells/mm2). Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, versus sham; ##p < 0.01, 60 versus 30 dpi. Scale bars, 100 μm. (C and D) In zebrafish, SA-β-gal+ cells co-localized with the senescence biomarker p21 and were devoid of the proliferation marker BrdU. Images were taken at 15 dpi. White arrows point to representative examples of co-localization, while yellow arrows point to the absence of co-localization. Scale bars, 100 μm. (E and F) In mice, a similar co-localization was found between SA-β-gal+ cells and the senescence biomarkers p16 and γH2AX. White arrows point to representative examples of co-localization. Scale bars, 100 μm. (G and H) At 15 dpi, the expression of senescence biomarkers, namely cdkn1a (encodes for p21), cdkn2ab (encodes for p15/16 in zebrafish), cdkn2a (encodes for p16 in mice), and p53, was evaluated by qPCR. Results are presented as relative expression toward sham. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, versus sham.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep.