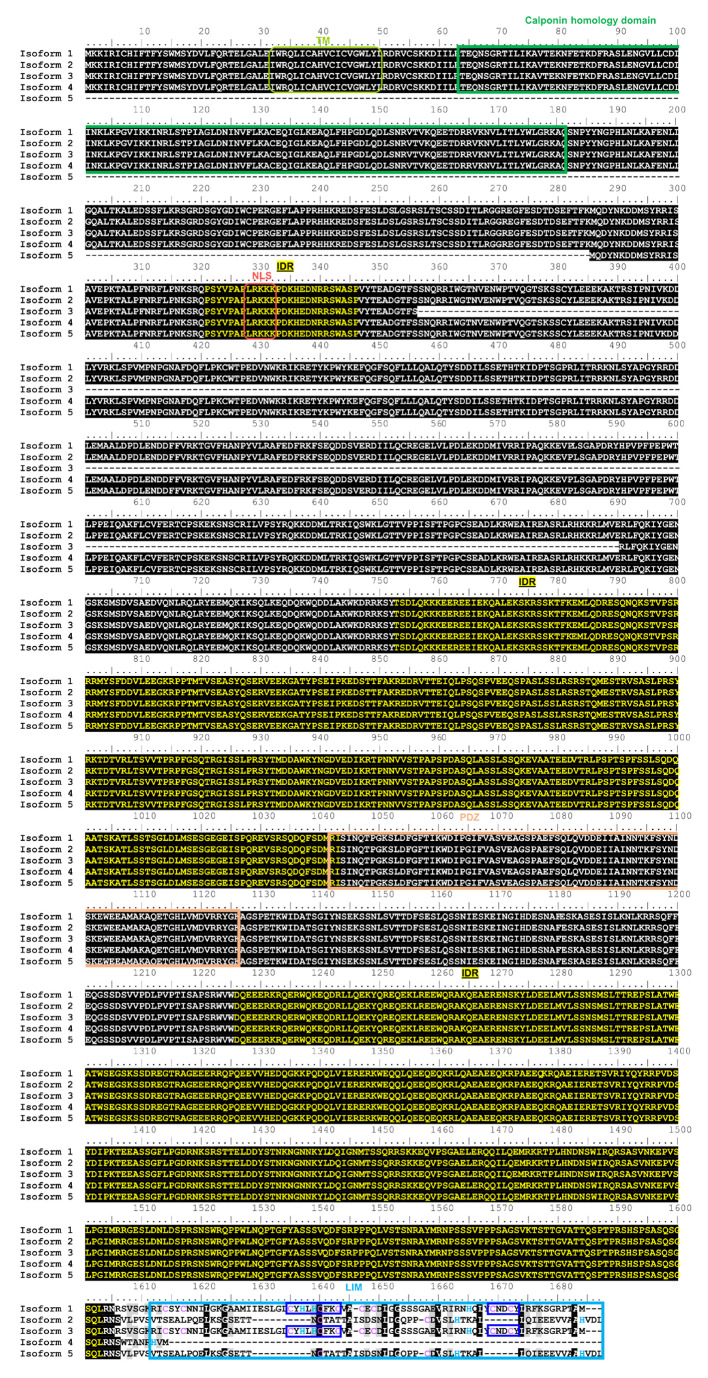
Figure 6 hLMO7 transcript variants show conserved intrinsic disorder but differ in the LIM domain. Alignment of hLMO7 isoforms 1 to 5 (UniProt IDs Q8WWI1-1; Q8WWI1-2; Q8WWI1-3; Q8WWI1-4; Q8WWI1-5, respectively) obtained by Clustal Omega [41]. The predicted domains, motifs and intrinsically disordered regions (IDR) reported in the Database of Disordered Protein Predictions (D2P2) [31] are marked by rectangles and the sequences indicated above. Isoform 5 lacks the putative transmembrane α-helix (TM, predicted by Phobius at https://phobius.sbc.su.se/) and the calponin homology domain (CH). Isoform 3 lacks residues 356–690. The LIM domain is missing in isoform 4. Isoforms 2 and 5 do not have the key zinc ion coordination motifs (dark blue rectangles). Cysteine and histidine residues that might also be involved in Zn(II) binding are marked in purple and blue, respectively. NLS, nuclear localization signal predicted by NLSdb (https://rostlab.org/services/nlsdb/).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Int. J. Mol. Sci.