Fig. 4
Fig. 4
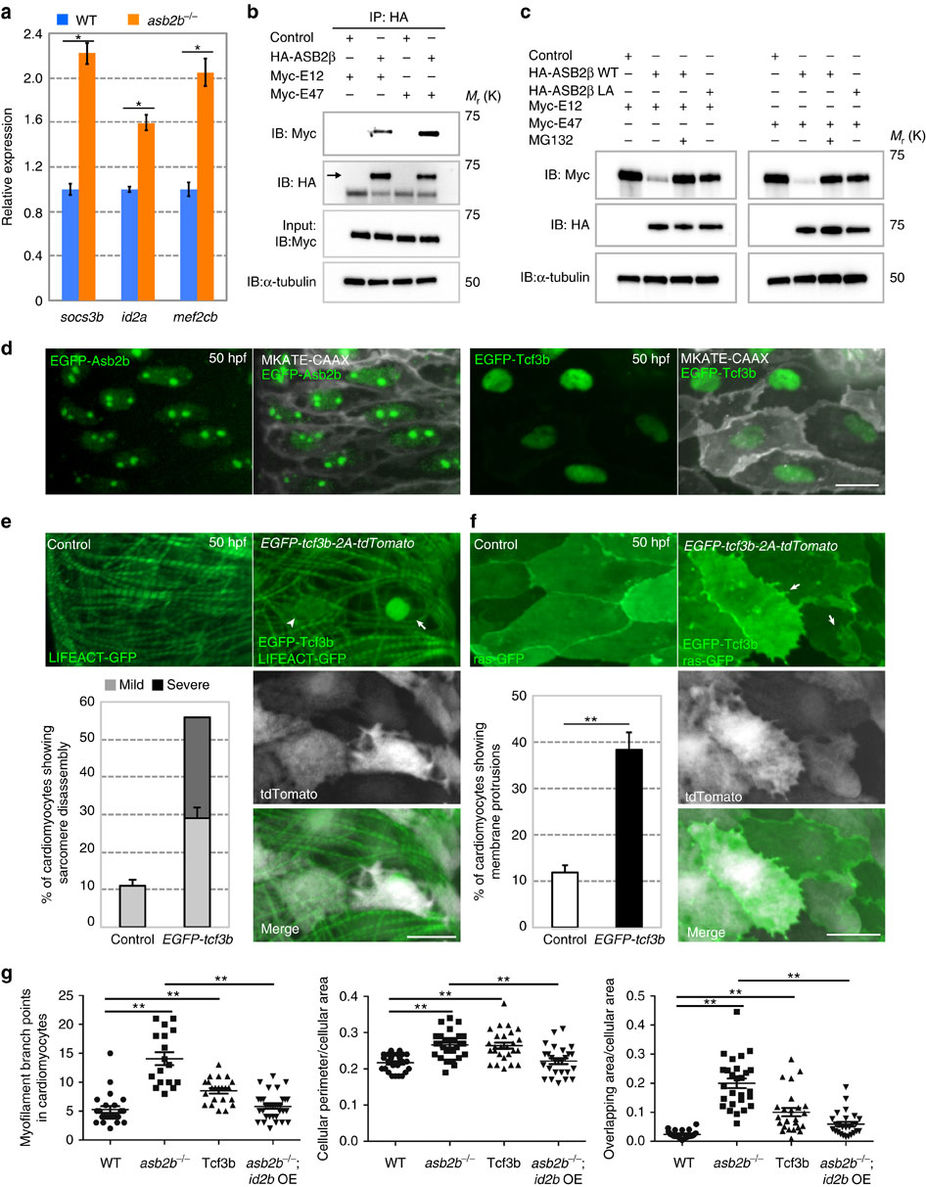
TCF3 is a target of ASB2 in cardiomyocytes.
(a) Relative mRNA expression (qPCR) of socs3b, id2a and mef2cb in isolated 50 hpf WT (blue bar) and asb2b mutant (orange bar) hearts (n=2 technical replicates; RNA samples obtained from 500 isolated hearts). (b) Co-immunoprecipitation (IP) of HA-ASB2β and Myc-E12 or Myc-E47 immunoblotted (IB) for HA (arrow) or Myc. Rat NCMs were transfected with HA-Asb2β or control (mock), together with Myc-E12 or Myc-E47 adenovirus vectors. All adenovirus vectors used contain an independent GFP cassette to assess transfection efficiency. After 24 h of transfection, cells were treated with 20 μM MG132 for 8 h and then IP was performed; lower bands in HA blot are HA antibody heavy chains. (c) Asb2β induced E12 and E47 degradation in a proteasome-dependent manner. Rat NCMs were transfected with control, HA-Asb2β or HA-Asb2β L595A mutant (Asb2βLA), together with Myc-E12 or Myc-E47 adenovirus vectors. After 48 h of transfection, NCMs were treated with 20 uM MG132 or DMSO for 12 h and these protein samples were extracted for immunoblotting. (d) Three-dimensional images of a 50 hpf TgBAC(asb2b:GFP-asb2b);Tg(myl7:MKATE-CAAX) atrium, and a Tg(myl7:MKATE-CAAX) atrium expressing EGFP-Tcf3b. Both Asb2b and Tcf3 localize to the nucleus in cardiomyocytes. (e) Three-dimensional images of 50 hpf Tg(myl7:LIFEACT-GFP) and Tg(myl7:LIFEACT-GFP);Tg(myl7:EGFP-tcf3b-2A-tdTomato) atria. (The transgenic fish exhibit mosaic expression of EGFP-tcf3b-2A-tdTomato in the heart.) EGFP-Tcf3b-2A-tdTomato expressing cardiomyocytes exhibit disassembly of myofilaments. Arrow points to a severe case of myofilament disassembly, arrowhead to a mild case. Number of cardiomyocytes exhibiting sarcomere disassembly (n=5 hearts, with averages taken from 20 cardiomyocytes per heart). (f) Three-dimensional images of 50 hpf Tg(myl7:ras-GFP) or Tg(myl7:ras-GFP);Tg(myl7:GFP-tcf3b-2A-tdTomato) atria. Tcf3b-2A-tdTomato expressing cardiomyocytes exhibit abnormal membrane protrusions (arrows). Number of cardiomyocytes exhibiting membrane protrusions (n=5 hearts, with averages taken from 20 cardiomyocytes per heart). (g) Quantification of myofilament branch points, cellular morphology and membrane protrusions in 50 hpf WT, asb2b mutant, Tg(myl7:GFP-tcf3b-2A-tdTomato) WT and Tg(myl7:id2b-2A-tdTomato) asb2b mutant atrial cardiomyocytes in Tg(myl7:LIFEACT-GFP) or Tg(myl7:ras-GFP) background. (n=15 to 34 cardiomyocytes from 3 to 6 hearts). *P<0.05 and **P<0.01 by one-way analysis of variance (ANOVA) followed by Tukey’s honest significant difference test. Error bars, s.e.m. Scale bars, 20 μm.