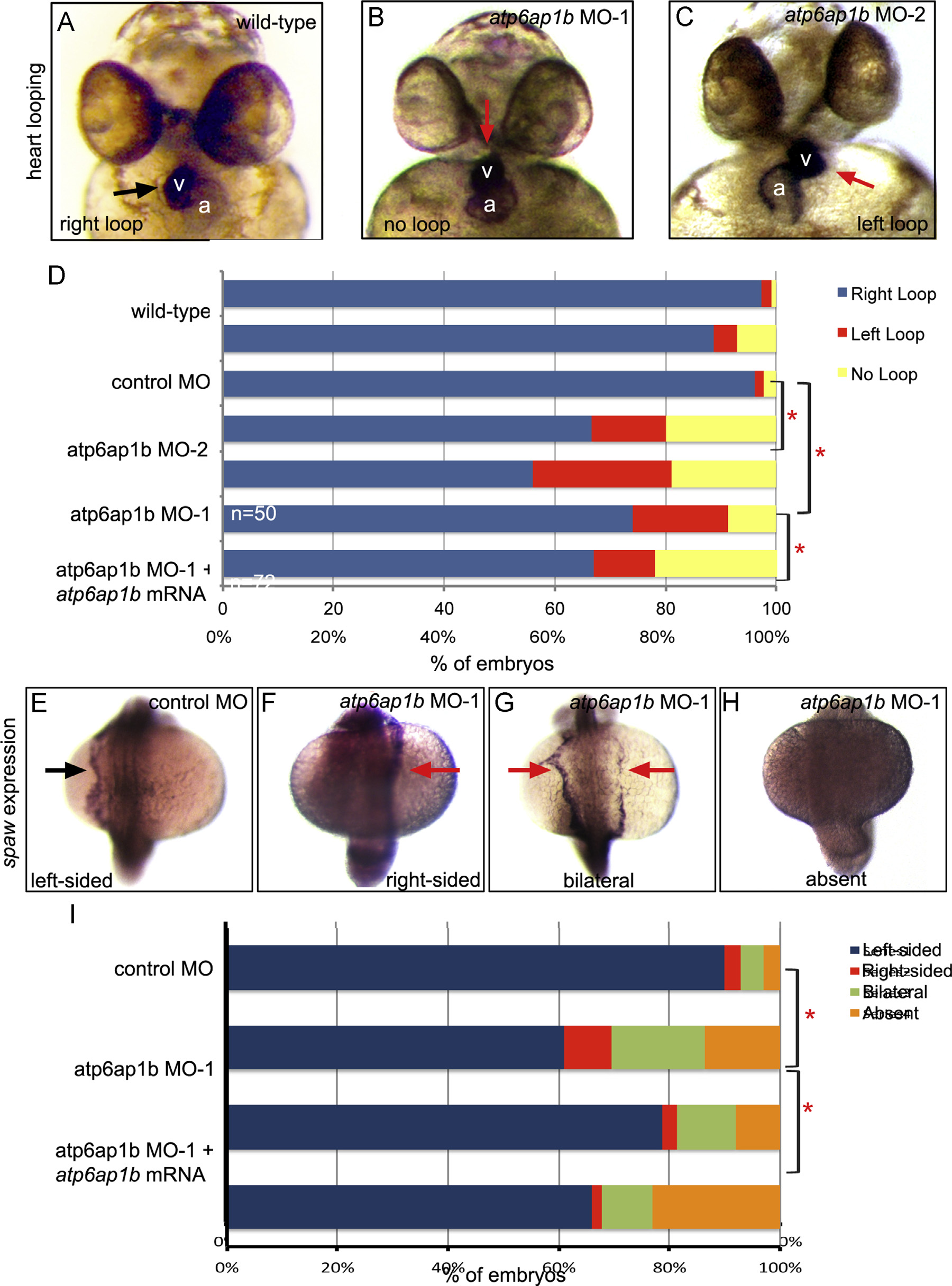
Fig. 3
Depletion of maternal and zygotic Atp6ap1b disrupted LR patterning. (A–C) RNA in situ hybridizations of the cardiac-specific cmlc2 marker were used to assess heart looping (arrow) at 2 dpf. v=ventricle; a=atrium. Normal rightward looping (A) was observed in wild-type embryos, whereas heart looping often failed (B) or was reversed (C) in embryos injected with two different translation-blocking MOs. (D) Quantification of heart looping phenotypes in atp6ap1b MO embryos. MO-induced heart looping defects were partially rescued by co-injection with atp6ap1b mRNA. (E–H) RNA in situ hybridization analysis of spaw expression (arrow) at 18 somite stage that is left-sided in controls (E), but reversed (F), bilateral (G) or absent (H) in many atp6ap1b MO embryos (I).
Reprinted from Developmental Biology, 407(1), Gokey, J.J., Dasgupta, A., Amack, J.D., The V-ATPase accessory protein Atp6ap1b mediates dorsal forerunner cell proliferation and left-right asymmetry in zebrafish, 115-30, Copyright (2015) with permission from Elsevier. Full text @ Dev. Biol.