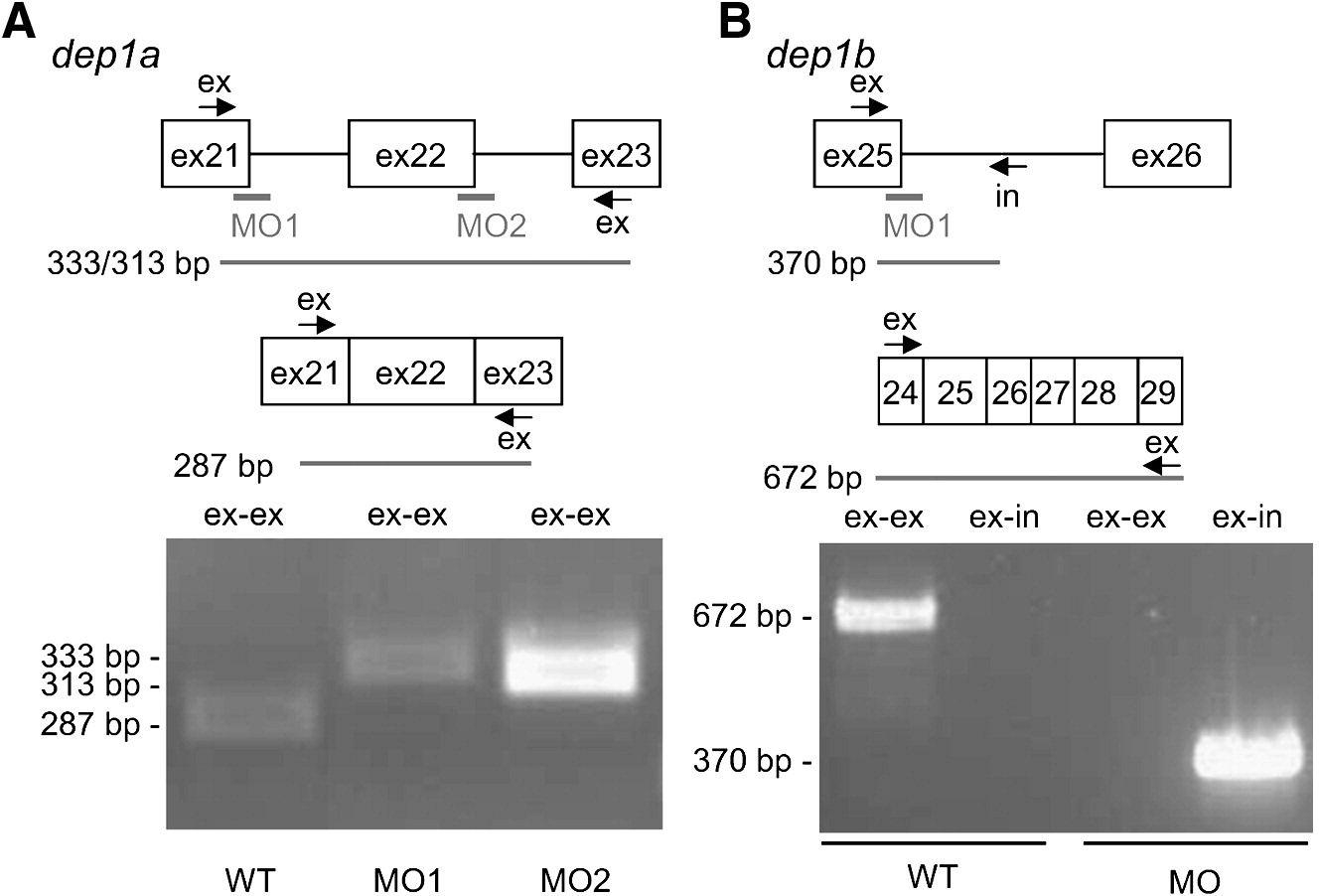
Fig. 2 Dep1a- and Dep1b-MOs efficiently blocked splicing. (A) Dep1a-MO1 and Dep1a-MO2 target the 3′ end of exon 21 and exon 22, respectively. Exon-specific primers were designed in exon 21 and exon 23, allowing PCR of spliced RNA (287 bp) and improperly spliced mRNA (333 bp and 313 bp), respectively. RT-PCR was done on non-injected control (WT), Dep1a-MO1 and Dep1a-MO2-injected embryos. The sizes of the obtained PCR fragments on an agarose gel are indicated. (B) The Dep1b-MO targets the 3′ end of exon 25, which encodes sequences corresponding to exon 21 of dep1a (see also Supplementary Fig. S1). Oligos were designed against exon 25 and intron 25, allowing PCR of unspliced dep1b RNA (370 bp) and as a control, exon-specific primers were used (672 bp). RT-PCR was done on RNA, isolated from non-injected control (WT) and Dep1b-MO-injected embryos and the resulting agarose gel is shown with the sizes of the bands indicated.
Reprinted from Developmental Biology, 324(1), Rodriguez, F., Vacaru, A., Overvoorde, J., and den Hertog, J., The receptor protein-tyrosine phosphatase, Dep1, acts in arterial/venous cell fate decisions in zebrafish development, 122-130, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.