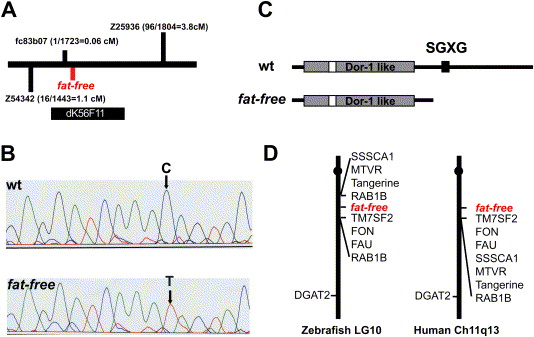
Fig. 3 Positional cloning of the zebrafish ffr locus A) Schematic of the ffr locus. Marker fc83b07 is 0.06 cM from the ffr locus (1 recombinant out of 1732 meioses). A radiolabeled probe synthesized from fc83b07 identified five BAC clones. Mapping using the end sequences of these BAC clones indicated that clone dK56F11 spanned the entire ffr locus. B) A single nucleotide mutation (C to T) that results in a premature stop codon was identified in ffr mutants (0 recombinants > 2000 meioses) within a predicted open reading of dK56F11. C) Domain analysis indicates that Ffr has a Dor-1 like domain, a tyrosine phosphorylation site (Tyr-P), and a SGXG motif. The truncated mutant protein is predicted to be approximately half the length of wild-type Ffr. D) ffr is located on zebrafish chromosome 10 in a region syntenic to a region of human chromosome 11q13.
Reprinted from Cell Metabolism, 3(4), Ho, S.Y., Lorent, K., Pack, M., and Farber, S.A., Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure, 289-300, Copyright (2006) with permission from Elsevier. Full text @ Cell Metab.