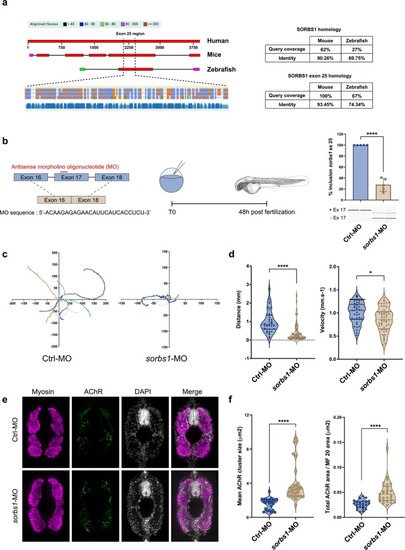
Exclusion of sorbs1 exon 17 during zebrafish development impairs locomotor phenotype. (a) Sequence alignment and domain mapping of the SORBS1 gene highlight a conserved region encompassing human exon 25, mouse exon 25 and zebrafish exon 17. This exon lies within the LIM domain‐containing region of the SORBS1 protein and exhibits high sequence conservation—with 93% identity between human and mice and 74% identity between human and zebrafish—underscoring its evolutionary conservation and functional relevance. Sequences from humans, mice and zebrafish have been aligned and compared using the Basic Local Alignment Search Tool (BLAST). (b) Schematic representation of the exon‐skipping strategy using antisense morpholino oligonucleotide (MO), and RT‐PCR analysis and quantification of sorbs1 exon 25 inclusion on total RNA extracts isolated from whole Ctrl‐MO and sorbs1‐MO embryos (48hpf); p < 0.0001, N = 5, unpaired Student's t‐test. Zebrafish eggs are microinjected with 0.6 mM of morpholino, and analysis is realized at 48 h post‐fertilization (hpf). (c) Colour‐coded video‐recorded tracks of swim trajectories of individual Ctrl‐MO and sorbs1‐MO embryos stimulated in touch‐evoked escape response (TEER) assay. (d) Violin plots representing the distribution of swimming distance and swimming velocity in TEER assay measured in cm, Ctrl‐MO (N = 37); sorbs1‐MO (N = 52) from three independent experiments p < 0.001 for distance, p < 0.05 for velocity, unpaired Student's t‐test. (e) Representative immunofluorescence of cryosectioned 48hpf zebrafish embryos. Images are confocal Z projections from Ctrl‐MO and sorbs1‐MO zebrafish. (f) Violin plots representing the distribution of AChR clusters size in mm2 and the distribution of AChR area normalized by skeletal muscle area in mm2; Ctrl‐MO (n = 23) from at least six fish; sorbs1‐MO (n = 29) from at least six fishes from three independent experiments. p < 0.001, unpaired Student's t‐test.
|

