Fig. 2
|
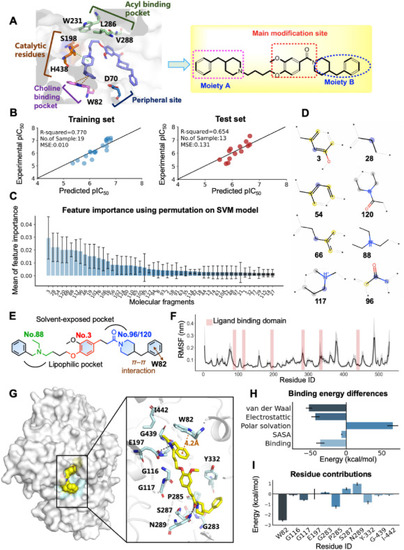
Computational chemistry revealed the binding modes of 8e. (A) Key residues of human BuChE (PDB code: 4TPK) and modification strategy for derivative design based on hit compound V1 structure. (B) The performance of the optimal QSAR model on the training and test dataset. Experimental vs. predicted –log10 (IC50) of the 19 training (left) and 13 test (right) samples. (C) Key molecular fragments derived from permutation feature importance. (D) Chemical structures of the top 8 fragments. (E) Key molecular fingerprints are shown in color in a 2D schematic. (F) The root mean square fluctuation (RMSF) of protein Cα during 50 ns MD simulations. Error bars represent standard deviation of three experiments. (G) The lowest energy binding mode of 8e–BuChE complex. The compound is shown in yellow stick mode. Key residues of BuChE are depicted in cyan stick mode. The hydrogen bond and π–π interactions were represented as dark and orange dashed line, respectively. (H) Total binding free energy and its component. (I) Residue contribution of hot residues for receptor–ligand combination. |