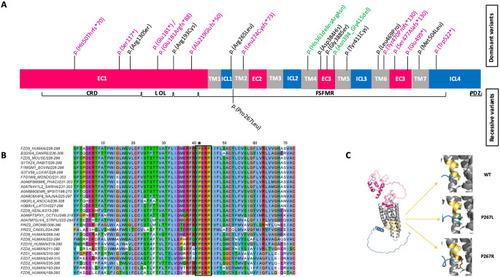
A novel recessive mutation in FZD5. (A) Likely pathogenic variants identified in FZD5. Truncating, missense, and in-frame variants are highlighted in violet, black, and green respectively. The upper section illustrates the position of the dominant (monoallelic) mutations identified in previous reports, while the lower section depicts the novel recessive (biallelic) mutation described in this study. FZD5 contains several important domains: a signal sequence at the N-terminus from amino acids (aa) 1–30; a conserved extracellular cysteine-rich Wnt-binding domain (CRD) from aa 31–150; a linker (L) from aa 151–193; an ordered loop (OL) from aa 194–227; and a Frizzled/Smoothened family membrane region (FSFMR) that includes seven transmembrane Frizzled domains (TM1: 228–260, TM2: 265–297, TM3: 314–345, TM4: 349–379, TM5: 396–428, TM6: 441–479, TM7: 499–522). A PDZ motif is located near the C-terminal at aa 583–585. (EC) extracellular domains and (C) cytoplasmic regions. (B) A multiple alignment was performed for 18 FZD5 orthologs from selected model species, along with all human Frizzled sequences. The YPERPI motif is highlighted (black rectangle); the p.Pro267Leu variant is indicated with an asterisk. Each sequence is identified by its UniProt accession number and the positions of its first and last amino acids in the selection. Amino acids color-coding follows the Clustal scheme. According to the alignment numbering, TM1 and TM2 comprise residues 3–34 and 42–72, respectively. Names of species are the following: DANRE, Danio rerio (Zebrafish); LOXAF, Loxodonta africana (African elephant); MONDO, Monodelphis domestica (Gray short-tailed opossum); PHACI, Phascolarctos cinereus (Koala); SARHA, Sarcophilus harrisii (Tasmanian devil); 9PSIT, Amazona collaria (yellow-billed parrot); NAJNA Naja naja (Indian cobra); ANOCA, Anolis carolinensis (Green anole, American chameleon); LATCH, Latimeria chalumnae (Coelacanth); XENLA, Xenopus laevis (African clawed frog); OCTVU, Octopus vulgaris (common octopus); STRPU, Strongylocentrotus purpuratus (Purple sea urchin); DROME, Drosophila melanogaster (Fruit fly); CAEEL, Caenorhabditis elegans. (C) (Left panel) Alphafold2 model of full-length human FZD5. The color code matches Fig. 1A. The YPERPI motif is highlighted and colored in yellow-orange. (Right panel) Zooming in on position 267 for the wild type (top), the P267L variant (middle), and the P267R variant (bottom). The two prolines of the YPERPI motif are depicted in yellow-orange sticks (P267 and P270), the leucine in deep teal sticks, and the arginine in chocolate sticks. Surrounding residues at position 267, including T339 from helix TM2, N350, and I353 from helix TM3, are shown in grey sticks
|

