- Title
-
Homozygosity for a hypomorphic mutation in frizzled class receptor 5 causes syndromic ocular coloboma with microcornea in humans
- Authors
- Cortés-González, V., Rodriguez-Morales, M., Ataliotis, P., Mayer, C., Plaisancié, J., Chassaing, N., Lee, H., Rozet, J.M., Cavodeassi, F., Fares Taie, L.
- Source
- Full text @ Hum. Genet.
|
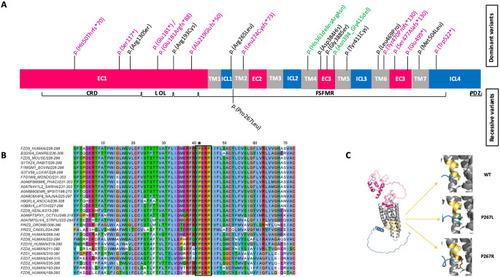
A novel recessive mutation in |
|
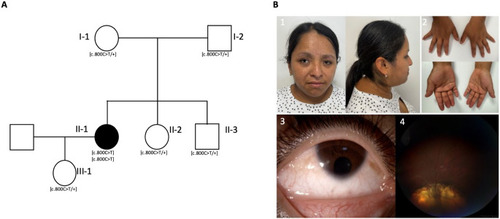
Clinical features associated with the c.800 C > T |
|
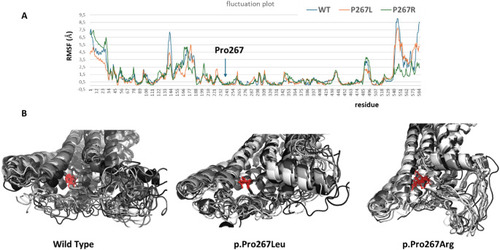
Fluctuation plot and molecular dynamics simulations ( |
|
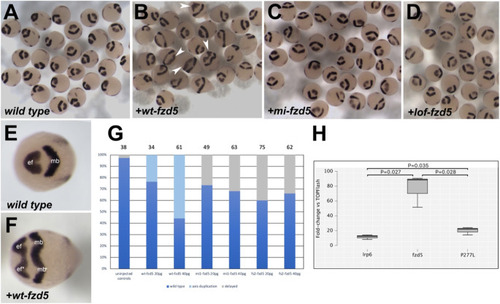
A missense variant in zebrafish Fzd5 mimicking the Pro267Leu variant does not induce a secondary neural axis. ( |
|
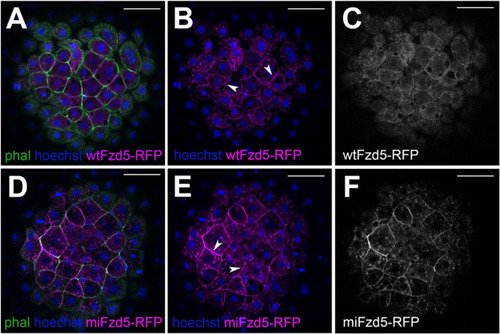
Subcellular localization of zebrafish Fzd5 is not affected by the Pro267Leu mutation. ( |