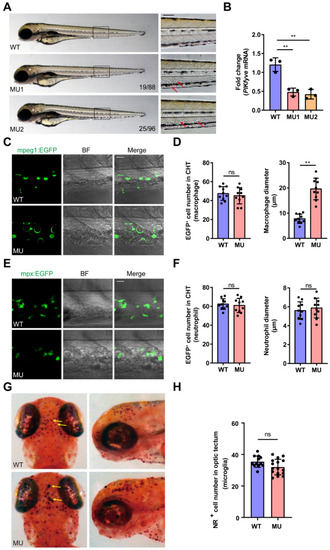
Defects in macrophages in PIKfyve mutant zebrafish embryos. (A) Representative images of WT and MU embryos at 3 dpf. The arrows show the cells with giant vacuole-like structures in the CHT. Cellular death in the mutant brains was evidenced by grey tissue (asterisk). (B) The relative expression levels of PIKfyve in the WT zebrafish and the homozygous mutants (MU1, MU2) were quantified by RT-qPCR. The data were analyzed by 2−∆∆CT method and ꞵ-actin was used as an internal control (** p < 0.01). (C–F) PIKfyve mutation was introduced into transgenic zebrfish lines in which macrophages (mpeg1:EGFP) (C,D) or neutrophils (mpx:EGFP) (E,F) were respectively highlighted with EGFP. In the mutant embryos, the macrophages were enlarged with giant vacuoles, while the neutrophils appeared normal. Scale bar, 20 µm. (D) Quantification of macrophage cell number (left panel) and macrophage diameter (right panel) in the CHT (ns, non-significant; ** p < 0.01). (F) Quantification of neutrophil cell number (left panel) and neutrophil diameter (right panel) in the CHT (ns, non-significant). (G,H) NR staining of microglia in WT siblings and PIKfyve mutants at 3 dpf. (G) Dorsal and lateral view of the microglia in the optic tectum of WT and MU larvae at 3 dpf stained with NR. The yellow arrows show the microglia stained with NR. (H) Quantification of NR-positive microglia cell number in the optic tectum of WT and MU larvae at 3 dpf (ns, non-significant).
|

