Fig. 4
- ID
- ZDB-FIG-240513-18
- Publication
- Qu et al., 2024 - Genome editing of FTR42 improves zebrafish survival against virus infection by enhancing IFN immunity
- Other Figures
- All Figure Page
- Back to All Figure Page
|
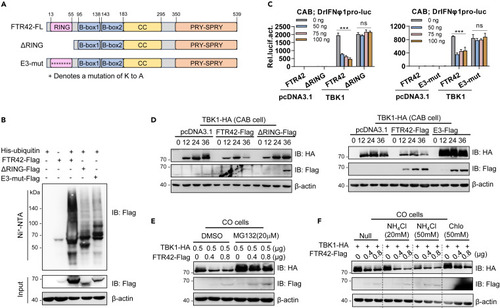
FTR42 inhibits TBK1-triggered IFN response dependent on its E3 ligase activity (A) Schematic diagrams of full-length FTR42, ΔRING and E3-mut. (B) FTR42 but not ΔRING or E3-mut possessed the E3 ligase activity. HEK293T cells seeded in 10 cm2 dishes were transfected with His-Ub, together with FTR42-Flag or ΔRING-Flag or E3-mut-Flag (5 μg each). 30 h later, the cells were lysed and incubated with Ni2-NTA overnight, followed by western blotting to determine the ubiquitination of FTR42 and two mutants. (C) FTR42-ΔRING or E3-mut failed to inhibit TBK1-triggered IFNφ1 promoter activation. CAB cells seeded in 48-well plates overnight were transfected with DrIFNφ1pro-luc, TBK1 or empty vector (100 ng each), together with FTR42 or ΔRING or E3-mut at increasing amounts. 30 h later, the cells were harvested for luciferase assays. (D) FTR42-ΔRING or E3-mut failed to attenuate TBK1 protein level in vitro. CAB cells seeded in 3.5 cm2 dishes were co-transfected with the indicated plasmids (1 μg each) for 30 h, followed by western blotting assays with tag-specific Abs. (E and F) FTR42-triggered TBK1 degradation was blocked by MG132 but not by NH4Cl or chloroquine. CO cells seeded in 12-well plates were co-transfected with indicated plasmids for 24 h, followed by treatment with MG132, NH4Cl, or chloroquine for additional 6 h. p values were calculated using Student’s t test. ∗∗∗p < 0.001; ns, not significant. |