Fig. 1
- ID
- ZDB-FIG-240513-15
- Publication
- Qu et al., 2024 - Genome editing of FTR42 improves zebrafish survival against virus infection by enhancing IFN immunity
- Other Figures
- All Figure Page
- Back to All Figure Page
|
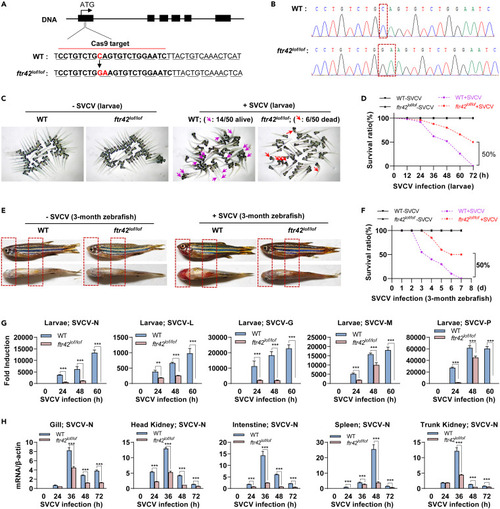
ftr42-deficient zebrafish show increased resistance to SVCV infection (A and B) Scheme of the genomic structure shows CRISPR/Cas9-targeted site in exon 1 of zebrafish FTR42, causing a reading frameshift due to 1-bp insertion and 1-bp mutation (A), which was verified by sequencing (B). (C) Representative images showed WT and ftr42lof/lof zebrafish larvae (4 dpf, n = 50 per group) immersed with SVCV (5×106 TCID50/mL) for 48h. Dead larvae showed obvious body curling. (D) Survival ratios of WT and ftr42lof/lof zebrafish larvae (n = 100 per group) were counted at indicated time points after immersion challenge with SVCV (5×106 TCID50/mL). (E) Representative images showed the hemorrhagic comparison of WT and ftr42lof/lof zebrafish adults (90 dpf) that were i.p. injected for 48h with SVCV (1×108 TCID50/mL), 20 μL per fish. (F) Survival ratios of WT and ftr42lof/lof zebrafish adults (n = 30 per group) were counted at indicated time points after injection challenge with SVCV (1×108 TCID50/mL). (G and H) FTR42 deficiency impaired SVCV replication in zebrafish larvae (G) and adults (H) following SVCV infection. WT and ftr42lof/lof zebrafish larvae (4 dpf) or adults (90 dpf) were challenged with SVCV as in (D) and (F), respectively. At indicated times, SVCV genes, including nucleoprotein (N), RNA polymerase (L), glycoprotein (G), matrix protein (M) and phosphoprotein (P) were detected by RT-qPCR. Zebrafish survival curves were analyzed by Log-Rank test. Data were shown as mean ± SD (N = 3). p values were calculated using Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001. |