Fig. 1
- ID
- ZDB-FIG-240513-1
- Publication
- Osman et al., 2024 - Non-invasive single cell aptasensing in live cells and animals
- Other Figures
- All Figure Page
- Back to All Figure Page
|
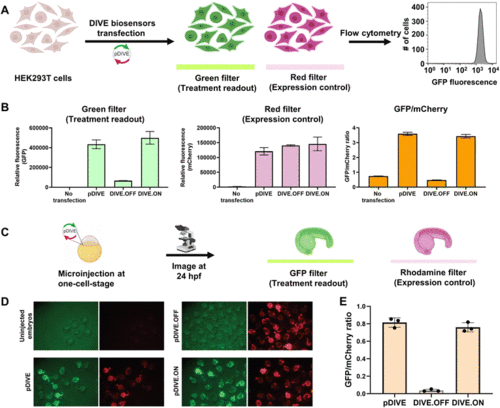
DIVE platform controls in HEK293T cells and zebrafish embryos. (A) Workflow of the cell assay. (B) Fluorescence measurements for each control: no transfection, the core plasmid pDIVE containing only GFP and mCherry, the DIVE.OFF negative control, and DIVE.ON positive control. GFP and mCherry fluorescence is measured for each cell population. The relative GFP/mCherry ratio is calculated and plotted. Data are the mean and standard deviation of three independent experiments each with technical triplicates. (C) The workflow for testing activity of the DIVE system in zebrafish embryos. WT embryos were injected at the one-cell stage and raised until 1 day post fertilization (dpf). (D) Epifluorescence micrographs from an entire clutch are shown for embryos. (E) Three random embryos from each injection condition were imaged and pixel intensity across the trunk quantified. Embryos lacking the mCherry expression control are not included as this indicates either they were not integrated into the genome or in a region with silent expression. The average GFP/mCherry ratio ± s.d. was plotted. Values for individual fish are shown. |