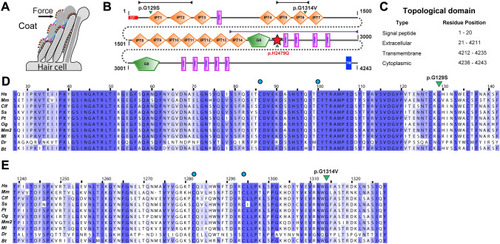
PKHD1L1 protein domain prediction and evolutionary analysis for missense variants (Family 1 and Family 3). a Schematic of a hair-cell stereocilia bundle under force stimulation highlighting the stereocilia surface coat. b Protein domain composition prediction from SMART using the Hs PKHD1L1 protein sequence as in NCBI accession code NP_803875.2, including the signal peptide (20 amino acids are predicted for Hs PKHD1L1 according to SMART. See Supplementary Table S1). Positions of each missense variant reported in this study are presented with a green arrowhead. The red star represents a newly predicted TMEM2-like domain. Black and purple arrow-headed lines represent the sequence fragments used for AlphaFold2 modeling of IPT1-2, IPT5-6, and TMEM2-like domain, respectively. c Topological description of Hs PKHD1L1 protein sequence as a reference. d, e Multiple protein sequence alignments comparing IPT1 and IPT6 domains among ten different PKHD1L1 orthologs, respectively (see Supplementary Table S1 for details about the selected species and Supplementary Fig. S3 for full PKHD1L1 sequence alignment). IPT1 has a pairwise sequence identity conservation of 82.3%, while IPT6 has a pairwise sequence identity of 74.9% across ten different orthologs. An independent % sequence identity analysis of only Hs and Mm species for IPT1 and IPT6 shows 82.9% and 77.8%, respectively (sequence alignment not shown). Missense variants are highlighted by green triangles. Blue circles represent cysteine residues forming disulfide bonds. Each alignment was color-coded for sequence similarity (35% threshold) using Jalview. White-colored residues report the lowest similarity and dark blue report the highest (see Methods). PKHD1L1 orthologs were chosen based on sequence availability and taxonomical diversity (Choudhary et al. 2020; De-la-Torre et al. 2018; Jaiganesh et al. 2018)
|

