Fig. 1
- ID
- ZDB-FIG-240112-35
- Publication
- Mahony et al., 2023 - Lineage skewing and genome instability underlie marrow failure in a zebrafish model of GATA2 deficiency
- Other Figures
- All Figure Page
- Back to All Figure Page
|
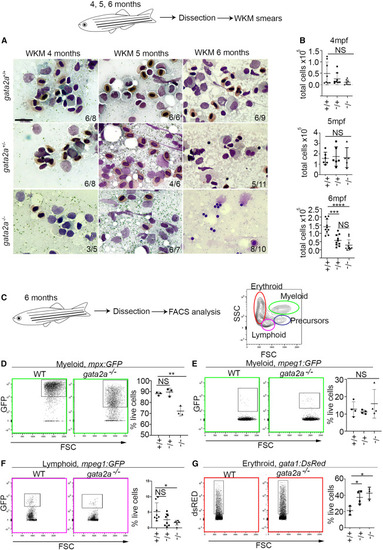
Loss of gata2a results in hypocellular marrow, neutropenia, B lymphopenia, and erythrocytosis at 6 mpf (A) WKM smears and May Grunwald/Giemsa staining at 4, 5, and 6 mpf. Scale bar, 10 μm. (B) Total WKM cell counts at 4 (n = 8/condition [biological replicates]), 5 (n = 6/condition [biological replicates] except gata2a−/−, where n = 5) and 6 mpf (n = 10/condition [biological replicates] except gata2a−/−, where n = 11). Error bars are mean ± SD. (C) Flow cytometry gating strategy. (D–G) Analysis of different lineages using either mpx:GFP, mpeg1:GFP, or gata1:dsRed transgenic lines. We examined neutrophils (mpx:GFP, myeloid gate),15 macrophages (mpeg1.1:GFP, myeloid gate), B cells (mpeg1.1:GFP, lymphoid gate),16 and erythrocytes (gata1:dsRed, erythroid gate)17 at 6 mpf. (D) gata2a+/+ vs. gata2a+/−: p = 0.82, gata2a+/+ vs. gata2a−/−: p = 0.0024, n = 3/condition (biological replicates). (E) gata2a+/+ vs. gata2a+/−: p = 0.87, gata2a+/+ vs. gata2a−/−: p = 0.64, n = 4/condition (biological replicates). (F) gata2a+/+ vs. gata2a+/−: p = 0.053, gata2a+/+ vs. gata2a−/−: p = 0.0145, n = 8/condition (biological replicates) except gata2a−/−, where n = 6. (G) gata2a+/+ vs. gata2a+/−: p = 0.0374, gata2a+/+ vs. gata2a−/−: p = 0.0146, n = 3/condition (biological replicates) except gata2a+/−, where n = 4. Analysis was completed using one-way ANOVA, not significant (NS) p > 0.05, ∗p < 0.05. (B and D–G) Each dot represents one sample. Data shown as mean ± SD. |