Fig. 2
|
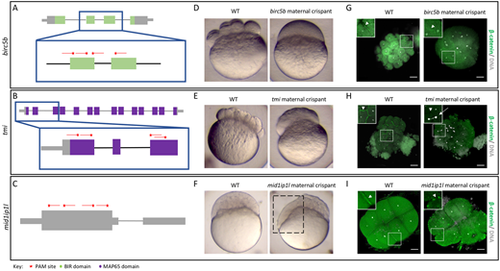
Identification of known maternal-effect genes using maternal crispants. (A-I) Known maternal-effect phenotypes resulting from the mutagenesis of three genes, birc5b (A,D,G), tmi (B,E,H) and mid1ip1l (C,F,I), were replicated using the maternal crispant technique. (A-C) Gene structure diagrams of birc5b (A), tmi (B) and mid1ip1l (C) showing the target sites of guide RNAs (red lines) and PAM sites (red stars). (D-F) Representative comparisons of a live wild-type (WT) embryo with live time-matched maternal crispant embryos of birc5b [D; 75 min post-fertilization (mpf)], tmi (E; 75 mpf) and midip1il (F; 150 mpf), showing defects in cytokinesis identical to previous studies. The mid1ip1l maternal crispant embryo shown exhibits a partially syncytial phenotype (box in F). (G-I) Immunohistochemistry labeling of β-catenin and DAPI staining in birc5b (G; 90 mpf), tmi (H; 90 mpf) and mid1ip1l (I; 75 mpf) maternal crispants showing the lack of accumulation of β-catenin in mature furrows, confirming a failure in furrow formation in spite of DNA replication. mid1ip1l maternal crispants contain ectopic β-catenin-containing vesicles near the cortex. In all immunohistochemistry labeling, insets show high magnification images of the boxed areas, highlighting the differences in β-catenin localization (arrowheads) and asymmetric DNA segregation (arrow). Note that there is some embryo size variation caused by methanol storage of embryos. Scale bars: 100 μm. |