Fig. 1
|
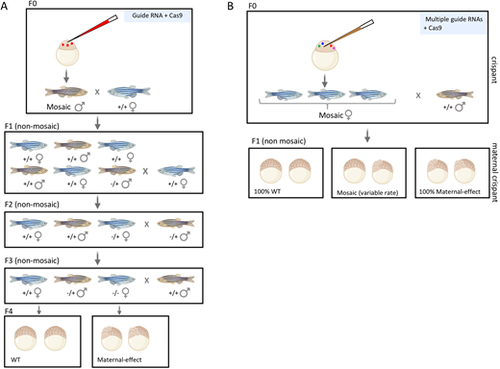
Maternal crispants can be used to efficiently identify maternal-effect genes in zebrafish in a single generation. (A) The traditional workflow for CRISPR-Cas9 in zebrafish uses one guide RNA that is co-injected with Cas9 mRNA or protein into the one-cell embryo to target a specific gene of interest. Successful targeting will result in adults with a mosaic germ line. To isolate these alleles, non-mosaic F1 fish are generated by crossing the F0 CRISPR-injected fish with a wild-type fish, then screening for heterozygous carriers of indels. Outcrossing followed by incrossing results in female homozygous mutants for a maternal-effect gene, phenotypes of which can be assessed in their offspring. (B) The maternal crispant technique generates maternal-effect phenotypes in a single generation. This strategy multiplexes four guide RNAs to target a single gene, thus increasing the rate of biallelic editing. If this biallelic editing event occurs in the germ line, the F1 offspring of F0 CRISPR-injected females have the potential to display a maternal-effect phenotype. |