Fig. 1
- ID
- ZDB-FIG-230912-15
- Publication
- Mathavarajah et al., 2023 - PML and PML-like exonucleases restrict retrotransposons in jawed vertebrates
- Other Figures
- All Figure Page
- Back to All Figure Page
|
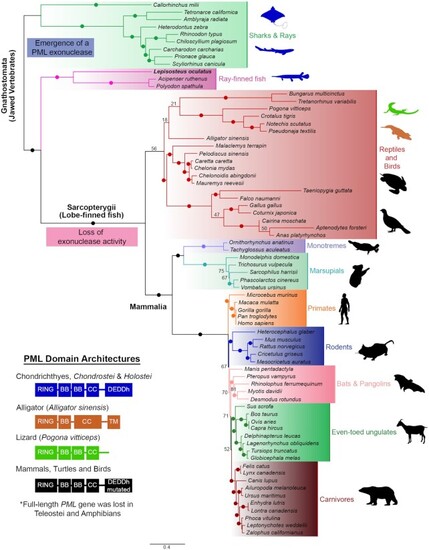
Domain architecture and Maximum Likelihood phylogenetic tree showing the relationships of PML homologs. 70+ protein sequences encoded in genomes from representative vertebrates were identified and utilized to construct the phylogenetic tree. PML emerged as an exonuclease in jawed vertebrates and eventually lost the catalytic residues for activity in amniote species. Teleost fishes and amphibians lack full-length PML-I. Ultrafast bootstrap values are shown as numbers if <85% and indicated with solid dots on branches if > 85%. Branch lengths indicate the expected number of amino acids substitution per site. Domains for homologs were determined using InterPro (https://www.ebi.ac.uk/interpro/) and four different domain architectures (left) were seen across analyzed sequences. Domain abbreviations refer to RBCC (Ring-B-box-Coiled Coil), BB (B-box), CC (Coiled coil), and DEDDh (3′-5′ DEDDh exonuclease). Sequences from species that were analyzed are shown in silhouettes and coloured according to the corresponding domain architecture on the left. Blue silhouettes indicate representative species with a PML ortholog encoding a predicted exonuclease domain. Silhouettes for species were obtained from PhyloPic (https://phylopic.org). |