FIGURE
Fig. 1
- ID
- ZDB-FIG-230905-11
- Publication
- Guo et al., 2022 - CRISPR/Cas9 screen in human iPSC-derived cortical neurons identifies NEK6 as a novel disease modifier of C9orf72 poly(PR) toxicity
- Other Figures
- All Figure Page
- Back to All Figure Page
Fig. 1
|
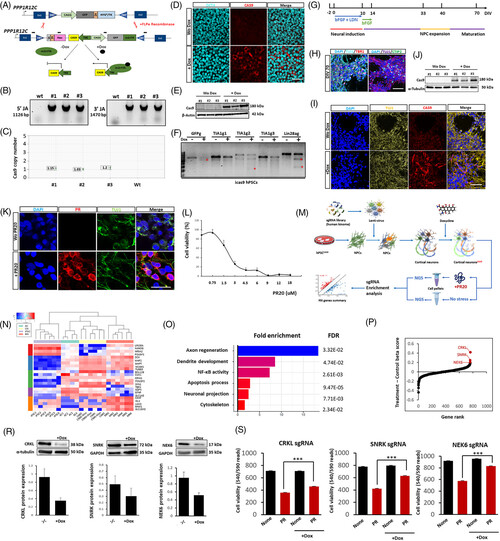
Kinome-wide CRISPR/Cas9 interference screen in doxycycline-inducible Cas9-iPSC-derived cortical neurons. (A) Schematic diagram of the strategy used to create inducible expression of Cas9 in iPSC lines: schematic representation of the AAVS1 locus, engineered to contain a flippase recognition target (FRTs)-flanked cassette suitable for recombinase-mediated cassette exchange (RMCE) using the pZ: F3-P TetOn-3×F-Cas9-F flanked by FRTs.17 This donor vector also contains a Tet-On system, including the M2 reverse tetracycline transactivator (M2rTA) and a tetracycline response element (TRE) upstream of a minimal cytomegalovirus (CMV) promoter. Neo: neomycin. (B) Identification of correctly targeted Cas9-iPSC cell clones by PCR, amplifying the 3′ and 5′ junctions (#1, #2, #3 denote the clone number, JA: Junction assay). (C) Digital droplet PCR (ddPCR) detection of copy number of CAS9 in the BJ1 iPSC line. (D) Immunostaining validation of Cas9 expression after doxycycline (3 μg/ml for 5 days) treatment in iPSCs. (E) Western blot validation of Cas9 expression after doxycycline treatment. (F) T7EI assay for Cas9-mediated cleavage in BJ1 Cas9-iPSCs using single sgRNAs targeting GFP (GFPg), T-cell-restricted intracellular antigen-1 (TIA1; TIA1g1, TIA1g2, TIA1g3), Lin-28 homolog A (LIN28A; LIN28Ag). (G) Schematic protocol of cortical neuron differentiation. LDN = LDN-193189. (H) Staining of DIV 80 PSC-cortical neuronal progeny for different cortical neuron markers (CTIP2 (or BCL11B) and T-box brain transcription factor 1 (TBR1)), neuron tubulin marker TUJ1 and DAPI. (I, J) Western blot and immunostaining for Cas9 expression in DIV80 PSC-neural progeny. (K) PR20 at a dosage of 6 μM was visualized by immunocytochemistry in DIV80 neural progeny treated with or without PR20, Scale bar = 20 μm. (L) PR20 dose-dependent cytotoxicity of DIV80 iPSC-neuronal progeny was measured by cell viability assay after a 24 hours treatment. (M) Schematic diagram of CRISPR/Cas9 screen procedure: DIV46 NPCs were transduced with the kinome-wide Brunello sgRNA vector library and transduced cells were selected with 1 μg/ml puromycin; NPCs were differentiated to DIV70 cortical neurons, after which 3 μg doxycycline was added for 5 days to induce Cas9, followed by treatment of 50% of the DIV 79 neuronal progeny with or without 6 μM PR20. The surviving cells were subjected to deep sequencing and statistical analysis for sgRNA distribution. Five biological replicates were performed. (N) RNA sequencing was performed on DIV80 iCas9 cortical neurons not treated with PR20, collected in parallel with the screen. Shown is expression of genes typically found in astrocytes (AC), cortical neurons (CN), iPSC and motor neurons (MN) (control iPSC, AC, and MN obtained from public available data with Gene Expression Omnibus (GEO) accession number: GSE98290). (O) Functional enrichment of candidate modifiers (113 genes that upon KO enhance survival under PR20 treatment) identified from the screen based on Gene Ontology Resource. (P) Hit gene distribution with normalization by negative control sgRNAs. (R) Western blot for NEK6, SNRK or CRKL in DIV80 neurons (normalized to α-Tubulin or GAPDH) following transduction with single sgRNAs against NEK6, SNRK, or CRKL, respectively. (S) Cell viability of Cas9-iPSC-derived DIV47 neurons transduced with single sgRNA guide against NEK6, SNRK, or CRKL with and without 6 μM PR20 treatment for 24 hours (n≥10). Data were plotted as mean ± SEM. Statistical significance was evaluated with one-way ANOVA and post-hoc Tukey's test; *,**,*** P values of <0.05, <0.01, <0.001 respectively. Scale bar = 50 μm
|
Expression Data
Expression Detail
Antibody Labeling
Phenotype Data
Phenotype Detail
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Alzheimers Dement