Figure 4
- ID
- ZDB-FIG-221214-208
- Publication
- Janoš et al., 2022 - Role of Monovalent Ions in the NKCC1 Inhibition Mechanism Revealed through Molecular Simulations
- Other Figures
- All Figure Page
- Back to All Figure Page
|
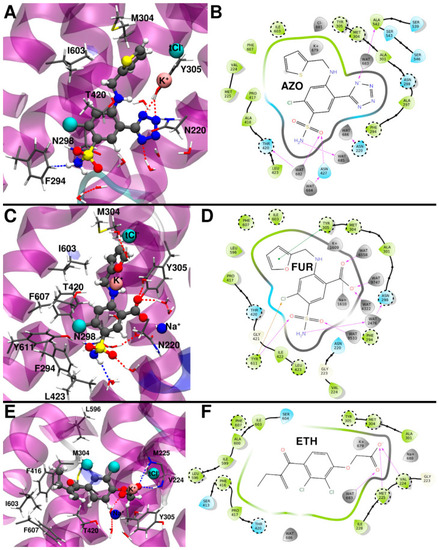
(A) Binding mode of azosemide (AZO) from the representative structure of the most populated cluster extracted from the molecular dynamics (MD) trajectory. (B) 2D schematic representation of the AZO/zNKCC1 interactions. (C) Binding mode of furosemide (FUR) from the representative structure of the most populated cluster extracted from the MD trajectory. (D) 2D schematic representation of the FUR/zNKCC1 interactions. (E) Binding mode of ethacrynic acid (ETH) from the representative structure of the most populated cluster from the MD trajectory. (F) 2D schematic representation of the ETH/zNKCC1 interactions. Polar/charged protein residues are colored blue, while the others are colored green. Waters and ions are colored gray. The most important protein residues, identified by alanine scanning analysis, are highlighted in dashed circles. |