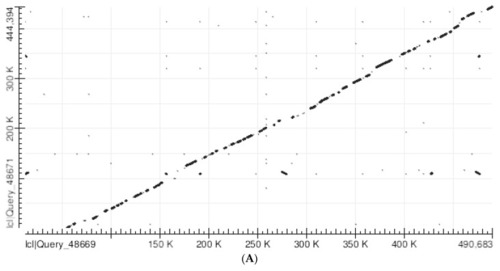
(A) Abscissa: Human SLC25A21 (490,793 nucleotides). Ordinate: Mouse slc25a21 (444,514 nucleotides). Number of matches: 1660. Note the significant alignment of most matches along the diagonal from the lower left corner to the upper right corner. The matches which are outside the diagonal mostly correspond to human and mouse TEs similar in structure. (B) Abscissa: Human SLC25A21 after the removal of all TEs. (298,209 nucleotides). Ordinate: Mouse slc25a21 after the removal of all TEs. (296,350 nucleotides). Number of matches: 266.The plot demonstrates a good overall alignment between the two sequences, while the gaps (which are usually short) correspond to poorly conserved sections. Note also the absence of significant matches outside the diagonal. (C) Abscissa: Human GHR (292,898 nucleotides). Ordinate: Mouse GHR (136,305 nucleotides). Number of matches: 280. Note the significant alignment of most matches along the diagonal from human GHR nucleotide 140,000 to the upper right corner, indicating that human nucleotides 1 to 140,000 have no matches with the mouse sequence, while the rest of the human sequence and the whole mouse sequence are homologous. The matches that are outside the diagonal mostly correspond to TEs that are similar in human and mouse. (D) Abscissa: Human GHR (homologous section only) after the removal of all TEs (85,058 nucleotides; from 78,000 to163,058). Ordinate: Mouse GHR after the removal of all TEs (89,777 nucleotides). Number of matches: 27. The plot demonstrates a general overall alignment between the two sequences, but the gaps are numerous and wide, corresponding to poorly conserved sections.
|

