Fig. 1
- ID
- ZDB-FIG-220620-12
- Publication
- Yao et al., 2022 - Comparing the folding landscapes of evolutionarily divergent procaspase-3
- Other Figures
- All Figure Page
- Back to All Figure Page
|
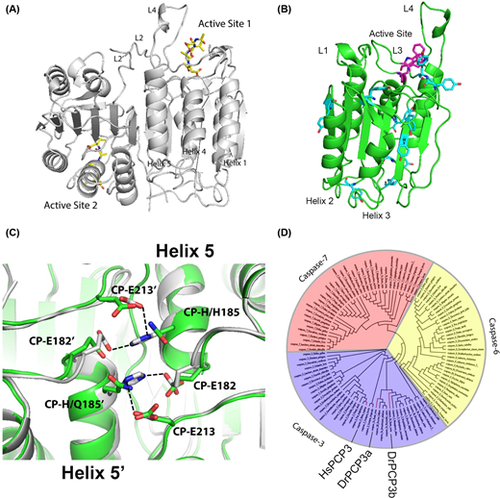
Caspase structure and phylogeny (A) Structure of human caspase-3 dimer is shown in gray (PDB ID: 1CP3). Inhibitors are shown in yellow, and active site loops L2, L2’ and L4 are labeled. (B) Caspase protomer with active site tryptophans (magenta) and tyrosines (cyan) labeled. Active site loops L1, L3 and L4 are shown, and helices 2 and 3 are labeled. (C) Comparison of contacts across the dimer interface (helix 5) for zebrafish caspase-3b (green) (modeled from human caspase-3) and human caspase-3 (gray) (PDB ID: 1CP3). CP refers to the common position numbering system as described in the text. Dashed lines show hydrogen bonds. (D) Phylogenetic tree of effector caspases. Caspase-3, -6 and -7 are marked in purple, yellow and red, respectively. Human caspase-3 (HsPCP3), zebrafish caspase-3a (DrPCP3a) and -3b (DrPCP3b) are labeled and shown by the red lines. |