Fig. 3
- ID
- ZDB-FIG-220305-25
- Publication
- Hoque et al., 2021 - Adaptation to an amoeba host drives selection of virulence-associated traits in Vibrio cholerae
- Other Figures
- All Figure Page
- Back to All Figure Page
|
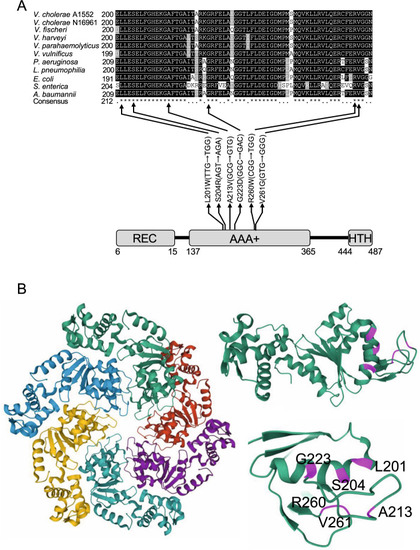
A The FlrA protein has three domains as indicated, an N-terminal signal receiver (REC) domain, a central ATPase-Associated domain with diverse cellular Activities (AAA+) and a C-terminal DNA binding helix turn helix (HTH) domain. Positions of the mutations in the central domain are indicated with respective amino acid and nucleotide base substitution in the codon. Affected amino acids in the sequence alignment of FlrA protein are indicated by black arrows. Protein sequences were retrieved from the NCBI protein database—V. cholerae A1552 (AUR70352), V. cholerae N16961 (NP_231768), V. fischeri (WP_011262363), V. parahaemolyticus (WP_025525752), V. vulnificus (WP_039545791), P. aeruginosa (NP_249788), L. pneumophila (WP_027221215), E. coli (MHO05571), S. enterica (WP_064013385), A. baumannii (SCY06189). B Structural model of FlrA generated from the crystal structure of the P. aeruginosa FleQ AAA+ domain is shown (5EXP). The monomers of the hexametric assemblies of the AAA+ domains of FleQ are shown in different colours (left). One monomer is shown with amino acid residues affected by nsSNPs highlighted in magenta (top right) and close-up view of the denoted residues is shown (bottom right). |