Fig. 4
- ID
- ZDB-FIG-220219-4
- Publication
- Seni-Silva et al., 2022 - Natterin-like depletion by CRISPR/Cas9 impairs zebrafish (Danio rerio) embryonic development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
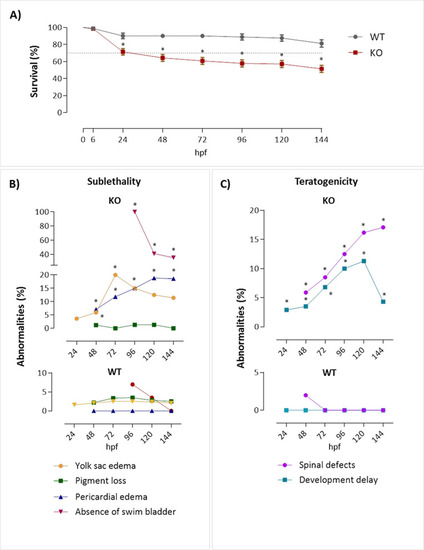
Survival rate, sublethal and teratogenic abnormalities of zebrafish after depletion of the |