Fig. 2
- ID
- ZDB-FIG-211118-65
- Publication
- Matrone et al., 2021 - Nuclear S-nitrosylation impacts tissue regeneration in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
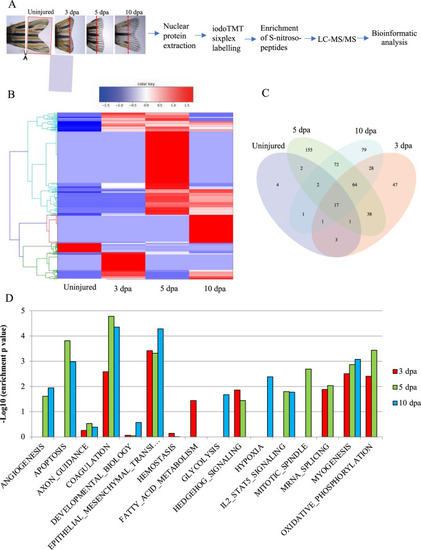
Bioinformatic analysis of the S-nitrosylome in zebrafish tailfin regeneration.
A Workflow for the analysis of the S-nitrosylome. Tailfins of zebrafish (6 months old) were amputated; regrown tissue was collected in uninjured and at 3, 5, and 10 dpa (dash red lines represent the edge of the amputation); nuclear proteins were extracted and labeled with iodoTMT, digested with trypsin, and S-nitroso-peptides enriched through anti-TMT antibody containing resin and followed by LC-MS-MS. B, C Hierarchical clustering heat map and Venn diagram showing the dynamic changes in the number of S-nitrosylated nuclear proteins during the regeneration compared to uninjured. D Hallmark pathways enrichment by the differentially expressed S-nitrosylated proteins during regeneration compared to uninjured. The significant pathways are displayed along the x-axis. Dpa day post-amputation. N = 2 biological replicates, followed by bioinformatic analysis. |