|
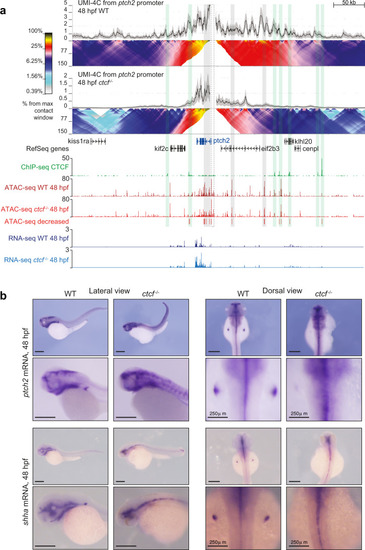
CTCF is required to sustain the regulatory landscape and expression pattern of the <italic>ptch2</italic> gene.a Top, UMI-4C assays in WT and ctcf−/− embryos at 48 hpf using the ptch2 gene promoter as viewpoint. Black lines and gray shadows represent the average normalized UMI counts and their standard deviation, respectively. Domainograms below UMI counts represent contact frequency between pairs of genomic regions. Bottom, tracks with CTCF ChIP-seq, ATAC-seq and RNA-seq at 48 hpf in WT and ctcf−/− embryos, as well as decreased ATAC-seq peaks in ctcf−/− embryos. A dotted-line square represents the restriction fragment containing the ptch2 gene promoter that is used as a viewpoint; green shadows highlight CTCF sites and gray shadows highlight downregulated ATAC-peaks without CTCF binding. Upregulated genes are shown in blue. b Whole-mount in situ hybridizations of the ptch2 and shha genes in WT and ctcf−/− embryos at 48 hpf. Left, lateral view; right, dorsal view. Scale bars represent 500 µm, unless indicated.
|