Figure 1.
- ID
- ZDB-FIG-210602-1
- Publication
- Chyżyńska et al., 2021 - Deep conservation of ribosome stall sites across RNA processing genes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
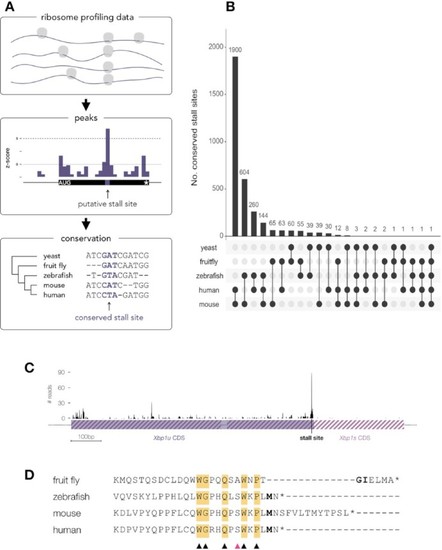
Conservation of stall sites across divergent eukaryotes. ( |