- Title
-
Deep conservation of ribosome stall sites across RNA processing genes
- Authors
- Chyżyńska, K., Labun, K., Jones, C., Grellscheid, S.N., Valen, E.
- Source
- Full text @ NAR Genom Bioinform
|
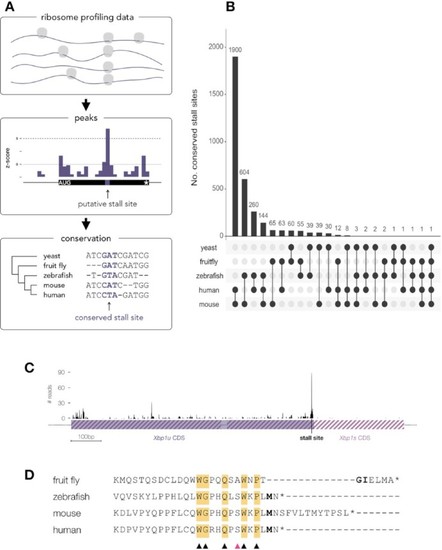
Conservation of stall sites across divergent eukaryotes. ( |
|
Mechanisms of conserved stalling. ( |
|
Functional characterization of CSS-containing genes. ( |