FIGURE 6
- ID
- ZDB-FIG-210310-29
- Publication
- Ranawakage et al., 2021 - Efficient CRISPR-Cas9-Mediated Knock-In of Composite Tags in Zebrafish Using Long ssDNA as a Donor
- Other Figures
- All Figure Page
- Back to All Figure Page
|
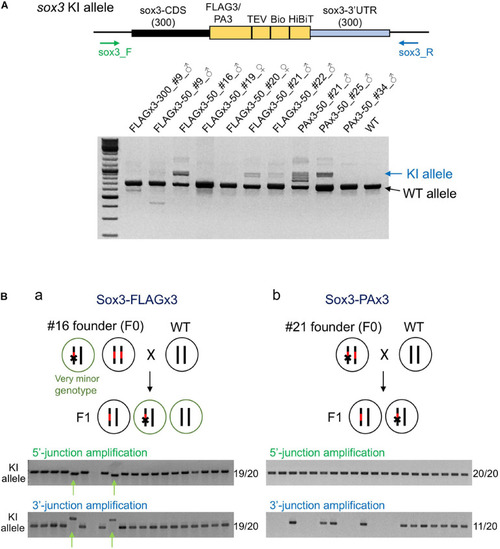
Founder germ cell mosaicism. |