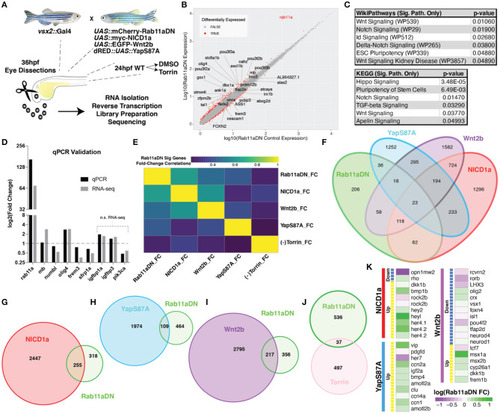
RNA-sequencing of Rab11aDN retinas identifies shared features with modulation of multiple signaling pathways control RPC proliferation. (A) Schematic of genetics for input for RNA-sequencing experiments to test changes in retinal transcript expression resulting from Rab11aDN expression, Notch-pathway activation (NICD1a), Wnt-pathway activation (Wnt2b), inactive Hippo pathway (YapS87A; Yap constitutive active), or inhibition of mTOR signaling (Torin). (B) Global analysis of differentially expressed transcripts in 36 hpf Rab11aDN retinas. Differentially expressed transcripts are indicated in Red. Gene names are listed for differentially expressed transcripts that display high residual to the mean. (C) Target pathways from IPA pathway analysis on Rab11aDN differentially expressed transcripts using WikiPathways (top) and KEGG pathways (bottom). (D) qRT-PCR validation of RNA-sequencing results for transcripts associated with the Notch (mb and numb), Hippo (olig4 and frem3), Wnt (sfrp1a), and mTOR (igfbp1a, igfbp3, pik3ca) pathway modulations. (E) Correlation of the fold changes of differentially expressed transcripts from Rab11aDN experiments with the fold changes observed in additional RNA-seq samples, indicating similarity between transcript signatures. (F–J) Venn diagrams of differentially expressed transcripts across RNA-sequencing samples of (F) All RNA-sequencing samples performed or pairwise comparisons of Rab11aDN differentially expressed transcripts with (G) NICD1a, (H) YapS87A, (I) Wnt2b overexpression studies, or (J) inhibition of mTOR signaling using the Torin inhibitor. (K) Heatmaps of Up- (Yellow) or Down-regulated transcripts within NICD1a, YapS87A, or Wnt2b experiments indicating the corresponding expression fold change in Rab11aDN studies.
|

