Fig. 6
- ID
- ZDB-FIG-210218-120
- Publication
- Kontur et al., 2020 - Ythdf m6A Readers Function Redundantly during Zebrafish Development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
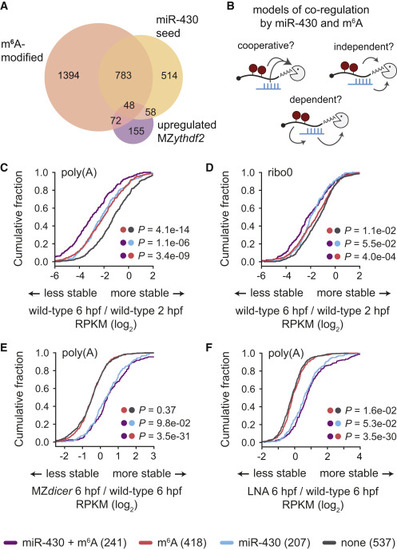
Figure 6. miR-430 and m6A Pathways Are Independent and Co-regulate Maternal mRNAs for Destabilization (A) Venn diagram of numbers of maternal mRNAs with a miR-430 seed in the 3′ UTR, are m6A modified, are stabilized in MZythdf2 (fold change > 0.5), or have overlapping features. (B) Schematic of potential mechanisms of miR-430 and m6A co-regulation, tested in (C)–(F). The pathways could function cooperatively, enhancing decay of common targets, independently, acting additively on common targets, or m6A could be dependent on miR-430, causing disruption of m6A-based mRNA decay upon loss of miR-430. (C–F) Cumulative distributions of fold changes in maternal mRNA abundance (log2 RPKM) for transcripts that are targets of both m6A and miR-430 (miR-430 + m6A, n = 241), m6A only (m6A, n = 418), miR-430 only (miR-430, n = 207), or neither (none, n = 537) from poly(A) (C, E, F) or ribo0 (D) mRNA-seq. Fold-change comparisons were between 6 and 2 hpf in wild-type (C, D) or at 6 hpf between MZdicer (E) or LNA-treated (F) embryos and wild-type controls. Dots indicate groups compared for corresponding p values, computed by a Mann-Whitney U test. Legend on bottom. |