Fig. 1
- ID
- ZDB-FIG-210205-11
- Publication
- Shen et al., 2018 - Enhancer Trapping and Annotation in Zebrafish Mediated with Sleeping Beauty, piggyBac and Tol2 Transposons
- Other Figures
- All Figure Page
- Back to All Figure Page
|
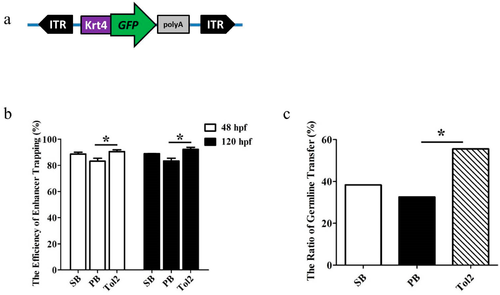
Green fluorescent protein (GFP) expression efficiency and germline transfer efficiency of enhancer trapping (ET) based on sleeping beauty (SB), piggyBa (PB) and Tol2 transposons in zebrafish. (a) Schematic of the transposon-based ET cassette (not to scale). The cassette includes two inverted repeat sequences (SB, PB, or Tol2 inverse terminal repeats (ITRs)), and a minimal Krt4 promoter, GFP, and β-globin polyA signal; (b) GFP expression efficiency was measured by GFP screen at 48 and 120 h postfertilization (hpf) after co-injection of a fixed amount of transposon plasmid with the corresponding transposase mRNA into zebrafish embryos. The total number of injected embryos was 1378 in SB, 1276 in PB, 1524 in Tol2. Error bars represent SD; (c) germline efficiency was measured by crossing the GFP-positive founders with wild-type (WT) fish. Significant difference was detected using a χ-squared test; asterisk indicates p < 0.05. |