Figure 3
- ID
- ZDB-FIG-210113-58
- Publication
- Kroll et al., 2021 - A simple and effective F0 knockout method for rapid screening of behaviour and other complex phenotypes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
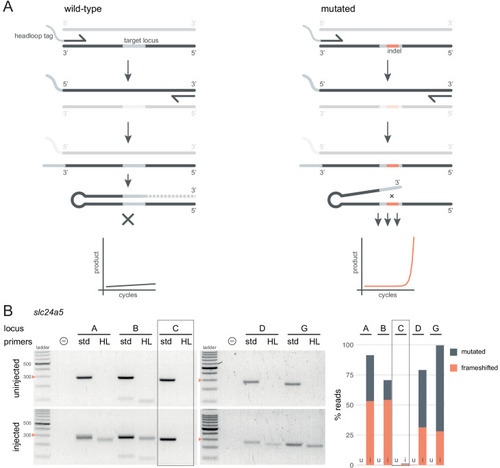
(A) Principle of headloop PCR. A headloop tag complementary to the target locus is added to one PCR primer (here, to the forward primer). During PCR, the first elongation incorporates the primer and its overhang; the second elongation synthesises the headloop tag. (left) If the template is wild-type, the complementary tag base-pairs with the target locus and directs elongation (hatched sequence). The amplicon forms a hairpin secondary structure, which prevents its subsequent use as template. (right) If the targeted locus is mutated, the tag is no longer complementary to the locus. The amplicon remains accessible as a template, leading to exponential PCR amplification. (B) (left) Target loci (A, B, C, D, G) of slc24a5 amplified with the PCR primers used for sequencing (std, standard) or when one is replaced by a headloop primer (HL). Orange arrowheads mark the 300 bp ladder band. (right) Deep sequencing results of the same samples as comparison (reproduced from Figure 2A, except locus C); u, uninjected control, i, injected embryo. Framed are results for gRNA C, which repeatedly failed to generate many mutations. See also Figure 3—figure supplement 1, 2, 3. |