Fig. 2
- ID
- ZDB-FIG-190821-39
- Publication
- Helker et al., 2019 - Whole organism small molecule screen identifies novel regulators of pancreatic endocrine development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
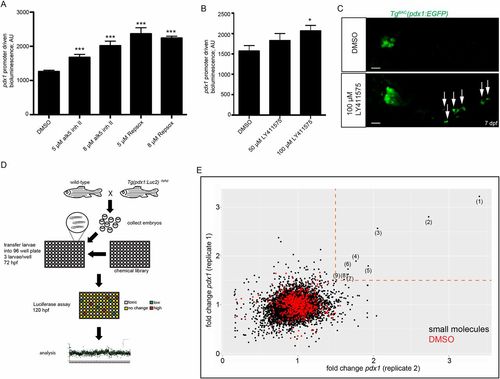
Whole organism screen for modulators of pdx1 expression. (A) Forty-eight-hour treatment of 72 hpf TgBAC(pdx1:Luc2) larvae with two TGFβ inhibitors (alk5 inh II and Repsox), which increase pdx1 mRNA levels in human islets, leads to an increase in luciferase activity. (B) Forty-eight-hour treatment of 72 hpf TgBAC(pdx1:Luc2) larvae with the γ-secretase inhibitor LY411575, which leads to increased differentiation of β-cells, increases luciferase activity. (C) Ninety-six-hour treatment of 72 hpf TgBAC(pdx1:EGFP) larvae with LY411575 leads to secondary islet formation as marked by TgBAC(pdx1:EGFP) expression (arrows). (D) Establishment of a high-throughput screening pipeline. Mating wild-type zebrafish with homozygous TgBAC(pdx1:Luc2)reporter zebrafish generates clutches of 100% hemizygous embryos. At 72 hpf, three larvae were transferred to each well of a 96-well plate and incubated for 48 h with a specific compound at 10 µM. At 120 hpf, each plate was incubated with long half-life luciferin and the bioluminescence intensity of each well measured in a standard plate reader. (E) Results of the primary screen. Differential pdx1 promoter-driven luciferase activity by small molecules of both duplicates is shown. A total of 8256 compounds from nine bioactive small molecule libraries were screened in duplicate. Each axis represents the levels of pdx1 reporter expression fold change from one of the duplicates. Dashed orange lines indicate 1.5-fold change. Red dots, negative control (1% DMSO); black dots, small molecules. (1) XL147; (2) SKF-86055; (3) chaulmoogric ethyl ester; (4) VPA; (5) HC toxin; (6) SB-736290; (7) PD168493; (8) scriptaid; (9) 2′,3′-O-isopropylideneadenosine. *P≤0.05, ***P≤0.001. Error bars represent s.e.m. Scale bar: 20 µm.
|